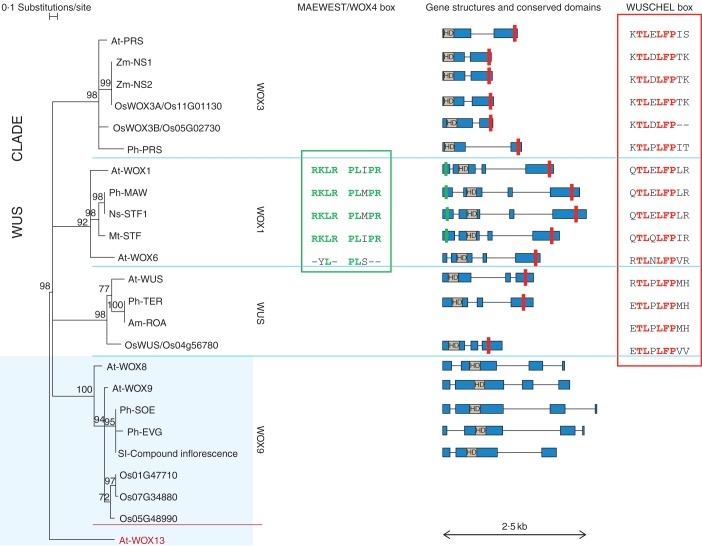
Fig. 1.
Phylogenetic tree of WOX sequences. WOX sequences from different species (Am, Antirrhinum majus; At, Arabidopsis thaliana; Mt, Medicago truncatula; Ns, Nicotiana sylvestris, Ph, Petunia × hybrida; Os, Oryza sativa; Sl, Solanum lycopersicum; Zm, Zea mays) are clustered into different subfamilies (WOX3, WOX1, WUS, WOX9) after phylogenetic analysis. The phylogenetic tree is based on the HOMEODOMAIN sequences. The WOX13 sequence (dark red) from arabidopsis is used as outgroup. To support WOX relationships, 1000 bootstrap samples were obtained using the software TREECON (Van de Peer and De Wachter, 1994). Bootstrap values <65 % are not shown and corresponding branches are displayed as unresolved. On the right of the tree, gene structure is displayed for most of the WOX gene sequences (solid blue bars are used for exons and thin black lines for introns), also depicting conserved amino acidic boxes (HD, HOMEODOMAIN; red rectangle, WUSCHEL box; green rectangle, MAEWEST/WOX4 box, as in Vandenbussche et al., 2009). In addition, conserved amino acid residues are displayed for the MAEWEST/WOX4 box and the WUSCHEL box. Note that WOX sequences also display characteristic C-terminal motifs with functional properties (not shown), as illustrated in Vandenbussche et al. (2009). Where not specified, accession numbers are the same as in Vandenbussche et al. (2009); Mt-STF (AEL30892.1), Nt-STF1 (AEL30893.1) and Sl-Compound Inflorescence (NP_001234072.1) are from GenBank.