Abstract
GPR22 is an orphan G protein-coupled receptor (GPCR). Since the ligand of the receptor is currently unknown, its biological function has not been investigated in depth. Many GPCRs and their intracellular effectors are targeted to cilia. Cilia are highly conserved eukaryotic microtubule-based organelles that protrude from the membrane of most mammalian cells. They are involved in a large variety of physiological processes and diseases. However, the details of the downstream pathways and mechanisms that maintain cilia length and structure are poorly understood. We show that morpholino knock down or overexpression of gpr22 led to defective left-right (LR) axis formation in the zebrafish embryo. Specifically, defective LR patterning included randomization of the left-specific lateral plate mesodermal genes (LPM) (lefty1, lefty2, southpaw and pitx2a), resulting in randomized cardiac looping. Furthermore, gpr22 inactivation in the Kupffer’s vesicle (KV) alone was still able to generate the phenotype, indicating that Gpr22 mainly regulates LR asymmetry through the KV. Analysis of the KV cilia by immunofluorescence and transmission electron microscopy (TEM), revealed that gpr22 knock down or overexpression resulted in changes of cilia length and structure. Further, we found that Gpr22 does not act upstream of the two cilia master regulators, Foxj1a and Rfx2. To conclude, our study characterized a novel player in the field of ciliogenesis.
Introduction
G-protein-coupled receptors (GPCRs) represent the largest protein superfamily in the human genome, including at least 800 receptors with a common seven-pass transmembrane domain organization. A large variety of ligands can bind and activate these receptors, such as light-sensitive compounds, pheromones, hormones, neurotransmitters, ions, peptides, proteins and many more. The endogenous ligands for more than 140 GPCRs remain however unidentified making them ‘orphan’ receptors [1], [2]. The current model of the receptor activation proposes that ligand binding induces a conformational change, changing the interaction with intracellular heterotrimeric G-proteins, consisting of a Gα and a Gβγ subunit. Subsequently, the receptor catalyzes the exchange of GDP to GTP in the Gα subunit. Next, the activated G protein subunits (α-GTP and βγ) regulate many downstream effectors, like adenylate cyclases, RhoGTPases and phospholipases [1]. In addition, GPCRs bind to β-arrestins and GPCR kinases (GRKs), which can activate G-protein independent signaling pathways, such as JNK, RhoA, MAPK or ERK [3]. Moreover, GRKs and β-arrestins are involved in GPCR desensitization by preventing G-protein coupling and stimulating endocytosis of the receptor [4], [5].
GPR22 is an orphan G-protein-coupled receptor, belonging to the class A rhodopsin-like GPCRs. The receptor was discovered together with three other human GPCR genes, using a customized search of a database of expressed sequence tags. RNA expression analysis revealed the presence of GPR22 in several human brain regions, such as the cerebellum, cerebral cortex and amygdale [6]. Furthermore, immunohistochemistry showed that GPR22 was present in cardiac myocytes and coronary arteries. Gpr22 null mice did not show any detectable phenotype, but had a significantly increased risk to functional cardiac decompensation following aortic banding. Overexpression of GPR22 in HEK-293 cells revealed that the receptor coupled to Gαi/Gαo, indicating that activation of the receptor would lead to the inhibition of adenylyl cyclase, a decreased production of cAMP and a decreased PKA activity [7]. Genome-wide association scan (GWAS) identified an osteoarthritis (OA) susceptibility locus on chromosome 7q22 that contains 6 genes, including GPR22 [8],[9]. In mice, GPR22 was expressed in the chondrocytes of the articular cartilage of knee joints after the induction of arthritis by papain or methylated bovine serum albumin and in the osteophytes of mice with instability-induced OA [8]. Nonetheless, the biological function and regulation of GPR22 remains poorly investigated and its role in the disease process is still debated [8]–[10].
GPCRs regulate many processes such as neuronal development, cardiac development, left-right (LR) patterning, cell division, inflammation and sensation (vision, taste, smell) [1], [11]. At the subcellular level, many GPCRs and their intracellular effectors are targeted to cilia [12]–[15]. Cilia are highly conserved eukaryotic organelles that protrude from the cell’s plasma membrane. They can be divided into 2 types, the primary immotile cilia and the motile cilia. Primary cilia are present on nearly every mammalian cell and are involved in the signal transduction of many pathways, involving HH, Wnt or GPCR pathways. Primary cilia consist of nine parallel outer microtubule doublets (‘9+0’). Motile cilia mostly have an additional central pair of microtubuli (‘9+2’), as well as radial spokes and dynein arms [16]–[18]. However, the zebrafish Kupffer’s vesicle also contains motile cilia that lack the central pair [16]. The main function of the motile cilium is to create a fluid flow, for example to move the fertilized egg into the oviduct, to circulate the cerebro-spinal fluid in the brain ventricles or to induce LR asymmetry in the embryo [17]–[19].
The zebrafish embryo is a commonly used in vivo model to study ciliogenesis. They develop ex utero, are transparent, easy to manipulate and develop many ciliated organs during early embryogenesis. Importantly, there are many well-characterized zebrafish ciliary mutants available. Typical characteristics of these ciliary mutants are a downwards curvature of the tail and LR patterning defects [20]–[22]. Zebrafish LR asymmetry is induced by a ciliated transient organ called the Kupffer’s vesicle (KV), which is similar to the mouse embryonic node. The KV is formed at early somitogenesis by the dorsal forerunner cells (DFCs) that form a ciliated epithelium around a fluid-filled lumen. The cilia inside the KV are motile and create a directional fluid flow directing asymmetric gene expression (for example southpaw, pitx2a, lefty 1/2) in the left lateral plate mesoderm (LPM). These genes induce the LR asymmetry of the developing organs, like the heart and the liver [23], [24]. Consequently, cilia defects often result in defective LR axis formation.
At the transcriptional level, two transcription factors act as master regulators of ciliogenesis, Foxj1 (forkhead box J1) and Rfx2 (regulatory factor X). They control the expression of genes encoding components necessary for cilia assembly or function, like dyneins, kinesins and tektins [25]. However, the details of the downstream pathways and mechanisms that maintain cilia length and structure are poorly investigated. The importance of identifying these regulators is underscored by many physiological processes and diseases, known as ciliopathies, in which cilia are involved.
In this study, we show that deregulation of gpr22 in the zebrafish embryo led to defective left-right (LR) patterning, caused by defects in KV cilia length and structure. Furthermore, our study indicates that Gpr22 does not act upstream of Foxj1a and Rfx2. Thus, Gpr22 is a novel player in the field of ciliogenesis, acting independently or downstream of the cilia master regulators.
Results
Deregulation of gpr22 results in a curvature of the tail and heart edema
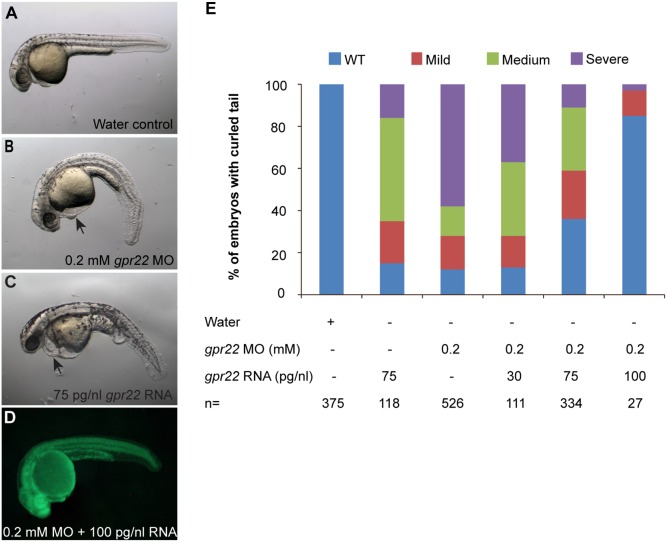
We cloned the zebrafish orthologue of human GPR22 (NCBI reference sequence zebrafish: NM_001044765.1; human: NM_005295.2). The amino acid sequence of zebrafish Gpr22 (NP_001038230.1) showed 78% overall identity to the human protein (NP_005286.2). The most conserved regions were found in the transmembrane domains (TM) (88% identity), the intracellular loops (IL) (84% identity) and the C-terminal tail (86% identity). Further comparison revealed conserved motifs reported to play a role in GPCR activation and signaling, like the DRY motif between TM3 and IL2 and the PxxY motif in the TM7 domain [26], [27]. To investigate the biological function of GPR22, we performed gain of function (GOF) and loss of function (LOF) studies in the zebrafish embryo. The LOF studies were carried out by injecting morpholino (MO) into one cell stage zebrafish embryos. As a control for potential off-target effects of the MO, we used two translation blocking MOs (a fluorescein tagged MO1 and a non-tagged MO2), each targeting different sequences at the 5′UTR of gpr22. Zebrafish gpr22 was first annotated as a single exon coding gene in the ZFIN, NCBI and Ensembl databases. Currently, the information provided by the databases about the splicing events of zebrafish gpr22 is inconsistent. Therefore, we decided not to use splicing blocking MOs. Injection with either of the MOs resulted in the same dose-dependent phenotypes, but MO1 appeared to be more efficient, inducing a phenotype at 0.2 mM instead of 0.4 mM (Fig. S1). Consequently, we used 0.2 mM of MO1 (further referred to as MO) for all subsequent experiments, unless otherwise specified. Since MO1 caused p53-mediated apoptosis in the head, a well-known off-target effect of MOs, all silencing experiments were performed in the presence of 0.1 mM of p53 MO (Fig. S1B, C) [28]. At 24 hpf, a knock down of gpr22 resulted in a downwards curvature of the tail, a reduction in length of the anterior/posterior (A/P) axis and heart edema. To ensure that the MO-induced tail phenotype was caused by a specific reduction of Gpr22 levels and not by off-target effects of the MO, we performed a rescue experiment by co-injecting fluorescein-tagged MO and capped, polyadenylated gpr22 mRNA into one cell stage zebrafish embryos. The green fluorescence of the MO allowed us to distinguish the rescued WT morphants from the WT embryos that were not efficiently injected with MO ( Fig. 1D ). Co-injection of gpr22 mRNA rescued the curly tail phenotype of the morphants in a dose-dependent manner ( Fig. 1 ). The GOF experiments were carried out by the injection of the same gpr22 mRNA into one cell stage zebrafish embryos. Injection of 75 pg/nl of gpr22 mRNA induced a curly tail phenotype, a reduction in length of the anterior/posterior (A/P) and heart edema at 24 hpf. The phenotype was dose-dependent ( Fig. 1 ; Fig. S2).
Figure 1. The downwards bending of the tail in gpr22 morphants is rescued by the co-injection of gpr22 mRNA.
(A–C) Lateral views with anterior to the left of zebrafish embryos at 24 hpf injected at one cell stage with (A) water, (B) 0.2 mM fluorescein tagged gpr22 MO, (C) 75 pg/nl gpr22 mRNA or (D) 0.2 mM fluorescein tagged gpr22 MO and 100 pg/nl gpr22 mRNA. (B, C) Deregulation of gpr22 results in a bending of the tail, a reduction in length of the anterior-posterior axis and heart edema (arrow). (E) Quantification of the tail phenotype. (D, E) The curly tail of embryos injected with fluorescein tagged gpr22 MO is rescued by the co-injection of gpr22 mRNA in a dose-dependent manner. MO = morpholino, hpf = hours post-fertilization, n = number of analyzed embryos, WT = wild type.
Deregulation of gpr22 results in defective LR patterning
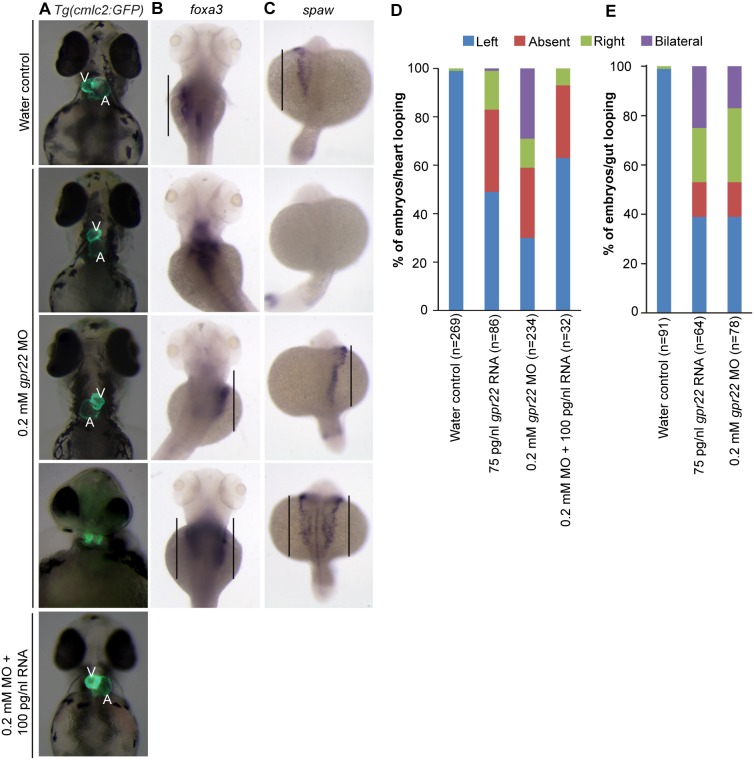
The downwards curvature of the tail and the heart edema are often associated with defects in ciliogenesis [22]. One of the most common consequences of defective ciliogenesis during development is a defective left-right (LR) asymmetry of the developing organs, e.g. the heart, liver or gut [22], [24]. To investigate the LR asymmetry of the heart, we injected gpr22 MO in transgenic zebrafish expressing green fluorescence protein (GFP) under the control of the cardiac myosin light chain 2 (cmlc2) promoter [29]. The LR asymmetry was defective in gpr22 morphants, which showed a normal, reversed, absent or bicardial cardiac looping at 4 dpf ( Fig. 2A, D ). Importantly, the heart phenotype could be rescued by the co-injection of gpr22 RNA ( Fig. 2A, D ). The position of the liver/gut was also randomized as visualized by whole mount in situ hybridization (WISH) for the liver/gut marker foxA3 ( Fig. 2B, E ). Finally, a WISH for the left-specific LPM markers southpaw (the zebrafish nodal), lefty1 and pitx2a at the 20 somite stage confirmed the randomized LR asymmetry in gpr22 morphants ( Fig. 2C ; Fig. S3; Table 1 ). Embryos with an overexpression of gpr22 also showed randomization of the left-specific LPM markers and consequently, a randomized cardiac looping and localization of the liver ( Fig. 2D, E ; Fig. S4; Table 1 ). These results suggest a possible function for Gpr22 in KV ciliogenesis.
Figure 2. gpr22 deregulation results in defective LR patterning.
(A) Ventral or (B, C) dorsal views with anterior to the top of embryos at (A, B) 4 dpf or (C) 20 somite stage. (A) Transgenic embryos expressing GFP under control of the cmlc2 promoter. In contrast to water controls, gpr22 morphants show (from top to bottom) normal, absent, reversed or bicardial cardiac looping. Co-injection of 0.2 mM gpr22 MO and 100 pg/nl gpr22 RNA partially rescues the randomized cardiac looping (last panel). (B) WISH for foxa3. The liver of water control embryos develops at the left side of the embryo. In contrast, the position of the liver in embryos injected with 0.2 mM gpr22 MO is randomized (straight line). (C) WISH for southpaw (spaw). gpr22 morphants show randomized expression of the early left-specific LPM marker southpaw (straight line). (D) Quantifications of the heart or (E) liver/gut phenotype. MO = morpholino, V = ventricle, A = atrium, dpf = days post-fertilization, n = number of analyzed embryos, GFP = green fluorescence protein, cmlc2 = cardiac myosin light chain type 2, LR = left-right, WISH = whole mount in situ hybridization, LPM = lateral plate mesoderm.
Table 1. Randomized expression of the left-specific LPM markers in zebrafish with deregulated gpr22 expression.
| spaw | lefty1 | |||||||||
| n | L (%) | R (%) | A (%) | B (%) | n | L (%) | R (%) | A (%) | B (%) | |
| Water control | 64 | 88 | 0 | 6 | 6 | 77 | 67 | 0 | 33 | 0 |
| 75 pg/nl gpr22 RNA | 24 | 42 | 4 | 50 | 4 | 83 | 50 | 7 | 40 | 3 |
| 0.2 mM gpr22 MO | 41 | 39 | 7 | 24 | 30 | 94 | 14 | 5 | 77 | 4 |
| pitx2a | ||||||||||
| n | L (%) | R (%) | A (%) | B (%) | ||||||
| Water control | 95 | 83 | 0 | 17 | 0 | |||||
| 75 pg/nl gpr22 RNA | 82 | 47 | 6 | 35 | 12 | |||||
| 0.2 mM gpr22 MO | 101 | 34 | 16 | 34 | 16 | |||||
MO = morpholino, n = number of analyzed embryos, L = left, R = right, A = absent, B = bilateral, LPM = left lateral plate mesoderm.
Gpr22 regulates LR asymmetry through its function in the KV
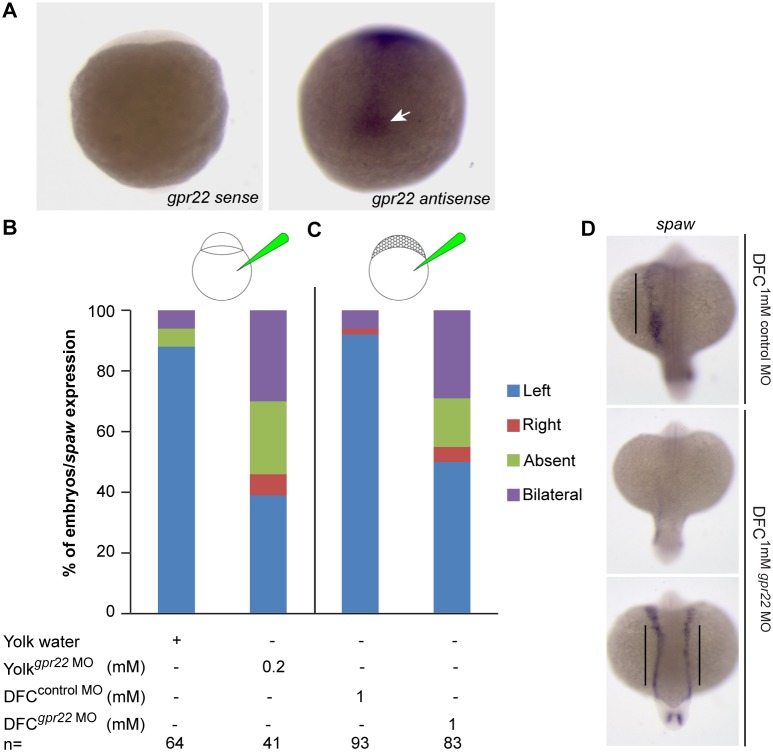
Induction of LR asymmetry in zebrafish depends on the rotational movement of the KV cilia to create a directional fluid flow [24], [30]. To investigate whether Gpr22 functions in the KV, we first analyzed the expression pattern of gpr22 during zebrafish and KV development, using two different gpr22 antisense probes and a control sense probe. During early stages of development, gpr22 was expressed ubiquitously (Fig. S5A). From bud stage onwards, the expression pattern became restricted to the axial structures and the developing KV ( Fig. 3A and Fig. S5B, B’). In contrast, we did not observe any signal in embryos treated with the control sense probe ( Fig. 3A ). The expression pattern of gpr22 at later stages of development (20 somite stage until long-pec) was previously described and reproduced by us (Fig. S5C–D’) (www.zfin.org) [31]. To investigate the KV-specific function of Gpr22 further, we performed a DFC/KV specific knock down of gpr22 by the injection of gpr22 MO into the yolk at the mid-blastula stage (3–4 hpf) ( Fig. 3C ), as previously described [29]. At this stage, the cytoplasmic bridges between the yolk and the embryo are closed, except for the ones between the yolk and the DFCs. Consequently, the MO diffuses only into the DFCs without affecting other embryonic cells. All the DFC/KV specific knock downs (injections at 1000-cell stage) were performed by injecting fluorescein-tagged control MO instead of water, or fluorescein-tagged gpr22 MO. The fluorescence allowed us to select embryos at 1–5 somite stage in which only the DFCs were hit [32]. The control MO was available in the lab and designed to target the 5′UTR of dus4l mRNA. However, injection of this MO in zebrafish embryos at one cell stage or mid-blastula stage did not induce phenotype and more specifically, did not cause left-right asymmetry defects ( Fig. 3C, D ). Therefore, we considered the MO as a valid control. Since the MOs often aggregated at the site of injection and did not diffuse into the DFCs, we increased the injected MO dose to 1 mM. A WISH for the LR marker southpaw at the 20 somite stage revealed that a KV-specific knock down of gpr22 induced randomized expression of southpaw, identical to the phenotype of a knock down in the whole embryo ( Fig. 3B–D ). These data indicate that Gpr22 regulates LR patterning through its specific function in the KV and not elsewhere.
Figure 3. Gpr22 regulates LR asymmetry through its function in the KV.
(A) WISH on 1–5 somite stage zebrafish embryos using the gpr22 sense or gpr22 antisense probe. Shown are the dorsal views of the tail bud region with anterior to the top. gpr22 is expressed in the axial structures and in the forming KV (arrow). (B, C) Percentage of embryos with randomized southpaw (spaw) expression, injected with (B) water or 0.2 mM gpr22 MO into the yolk at one cell stage (knock down in the whole embryo), or with (C) 1 mM control MO or 1 mM gpr22 MO into the yolk at mid-blastula stage (DFC/KV specific knock down). A DFC/KV specific knock down of gpr22 results in a similar percentage of zebrafish embryos showing randomized southpaw expression as a knock down in the whole embryo. (D) Representative images of panel (C). MO = morpholino, n = number of analyzed embryos, yolksuperscript = injected at one cell stage, DFCsuperscript = injected at mid-blastula stage, WISH = whole-mount in situ hybridization, DFC = dorsal forerunner cell, LR = left-right, KV = Kupffer’s vesicle.
Gpr22 affects cilia length and structure in the KV
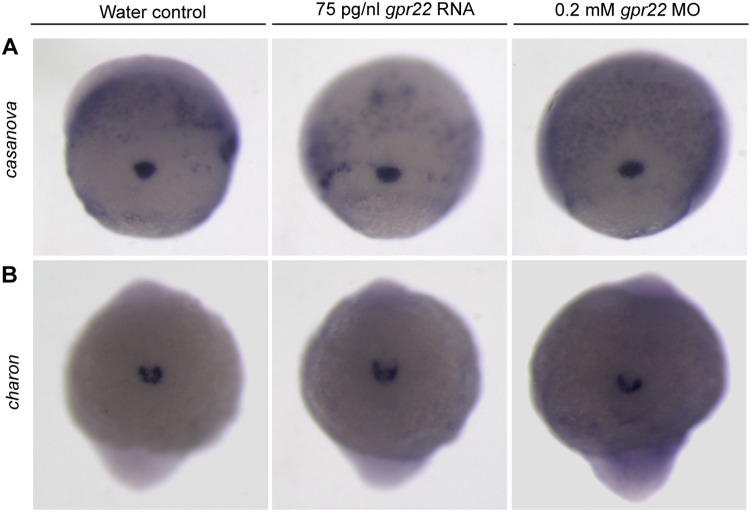
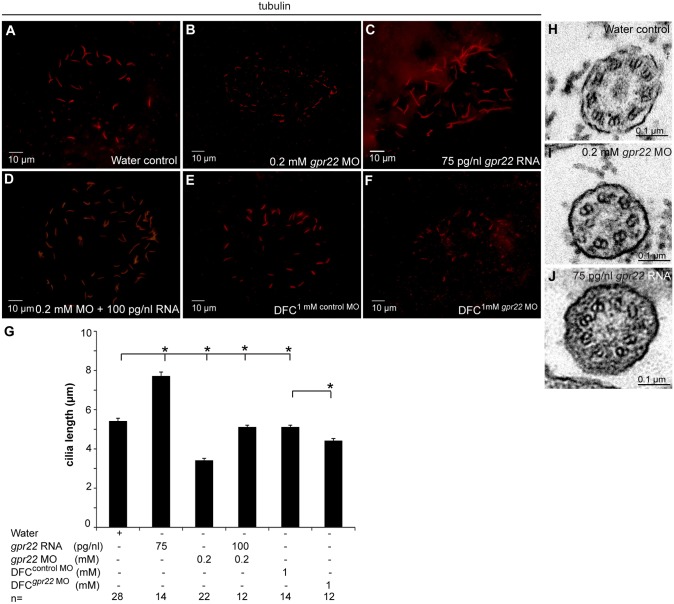
Since it is possible that gpr22 deregulation affects KV formation or morphology, we investigated the early steps of KV formation. It has been shown that the assembly of the DFCs is essential in the formation of a functional KV [33]. The DFC were visualized by a WISH for the marker casanova at 75%–85% epiboly [34],[35]. However, the assembly of the DFCs was not affected in embryos injected with gpr22 mRNA or MO ( Fig. 4A ). Next, we investigated KV morphology using a WISH for charon at the 10 somite stage. charon is expressed in the cells lining the KV and is a commonly used marker to study KV morphology [36], [37]. charon expression was not altered in embryos with deregulated gpr22 expression ( Fig. 4B ). These data support our notion that Gpr22 is not involved in KV formation or morphology. Another possibility is that Gpr22 is involved in KV ciliogenesis. It has been shown that the disruption of KV cilia number, length or structure leads to a loss of the directional fluid flow, a disturbed expression of the left-specific LPM genes and consequently defective LR patterning [24], [30]. Immunostaining with anti-acetylated tubulin antibody at 10 somite stage revealed that gpr22 morphants had significantly shorter KV cilia, compared to water injected controls (3.4±0.12 µm instead of 5.4±0.16 µm; P = <0.01) ( Fig. 5A, B, G ). In contrast, overexpression of gpr22 resulted in significantly longer cilia (7.7±0.22 µm instead of 5.4±0.16 µm; P = <0.01) ( Fig. 5: A, C, G). Moreover, the co-injection of 0.2 mM gpr22 MO and 100 pg/nl gpr22 RNA at one cell stage almost completely rescued cilia length (5.1±0.11 µm instead of 5.4±0.16 µm; P = <0.01) ( Fig. 5D, G ). Importantly, a DFC-specific knock down of gpr22 also resulted in significantly shorter cilia, compared to embryos injected at mid-blastula stage with control MO (4.4±0.13 µm instead of 5.1±0.11 µm; P = <0.01) ( Fig. 5E–G ). We noticed that the injection of the control MO at mid-blastula stage caused a very mild reduction in cilia length, compared to the injection of water at one cell stage (5.1±0.11 µm instead of 5.4±0.16 µm; P = <0.01) ( Fig. 5A, E, G ). However, this had no consequence on the expression of southpaw and did not induce LR asymmetry defects ( Fig. 3C, D ). We did not observe a detectable change in cilia number in all conditions tested. Next, we investigated whether the structure of the KV cilia was affected in embryos with deregulated gpr22 expression. Therefore, we performed a TEM ultra-structural analysis on cross sections of the KV cilia at 10 somite stage. Control cilia showed the typical 9 parallel outer microtubule doublets ( Fig. 5H ). In contrast, gpr22 overexpression or knock down resulted in a disruption of the proper microtubule arrangement or even the absence of doublets ( Fig. 5I, J ). Our data suggest that deregulation of gpr22 expression results in defective KV cilia length and structure, leading to LR asymmetry defects.
Figure 4. Gpr22 is not involved in KV formation or morphology.
(A) WISH for the DFC marker casanova on embryos at 75% epiboly or (B) for the KV marker charon on 10 somite stage embryos. Shown is (A) the dorsal view (B) of the tailbud region with anterior to the bottom. gpr22 overexpression or knock down does not alter the expression of casanova or charon. MO = morpholino, KV = Kupffer’s vesicle, WISH = whole mount in situ hybridization, DFCs = dorsal forerunner cells.
Figure 5. Gpr22 affects cilia length and structure in the KV.
(A–F) Immunostaining with anti-acetylated tubulin antibody on embryos at 10 somite stage injected at (A–D) one cell stage with (A) water, (B) 0.2 mM gpr22 MO, (C) 75 pg/nl gpr22 RNA, (D) 0.2 mM gpr22 MO and 100 pg/nl gpr22 RNA, or injected at (E, F) mid-blastula stage with (E) 1 mM control MO, (F) 1 mM gpr22 MO. Shown are the cilia of the KV. (G) Quantification of average cilia length ± s.d. (B, G) Whole embryo or (F, G) DFC-specific knock down of gpr22 significantly reduces cilia length. (C, G) In contrast, gpr22 overexpression results in significantly longer cilia. (D, G) Co-injection of 0.2 mM MO and 100 pg/nl RNA almost completely rescues cilia length. (H–J) TEM of cross sections of the KV cilia at 10 somite stage. (H) Structure of water control KV cilia showing 9 parallel outer microtubule doublets. (I) gpr22 knock down or (J) overexpression results in a disruption of the proper microtubuli arrangement or (I) even an absence of doublets. MO = morpholino, n = number of analyzed embryos, DFC = dorsal forerunner cells, DFCsuperscript = injected at mid-blastula stage, KV = Kupffer’s vesicle, TEM = transmission electron microscopy, * = P<0.01 (unpaired, two-tailed T-test).
Gpr22 does not act upstream of the cilia master regulators Foxj1a and Rfx2
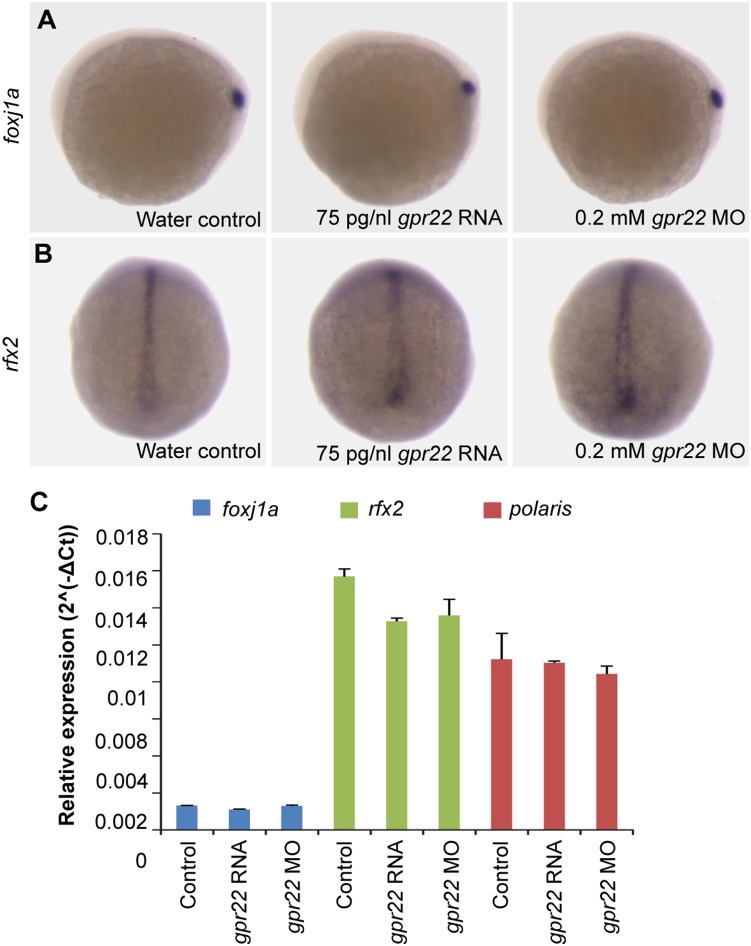
To investigate whether Gpr22 affects KV ciliogenesis by regulating the expression of foxj1a or rfx2, we carried out WISH and qPCR for these transcription factors on bud stage embryos. There were no detectable differences in the expression pattern or level ( Fig. 6A, B ) of these genes in LOF or GOF embryos. Furthermore, the expression level of a foxj1a/rfx2 target gene, polaris, was also unchanged, indicating that changes in gpr22 expression did not result in changes of the transcriptional activity of foxj1a or rfx2 ( Fig. 6C ). Thus, Gpr22 does not affect KV ciliogenesis by changing the expression or transcriptional activity of foxj1a or rfx2.
Figure 6. Gpr22 does not act upstream of Foxj1a/Rfx2 in KV ciliogenesis.
(A, B) WISH of (A) foxj1a or (B) rfx2 on embryos at bud stage. Shown are (A) the lateral view with anterior to the left or (B) dorsal view with anterior to the top. gpr22 knock down or overexpression does not alter the DFC expression of foxj1a or rfx2, two master regulators of ciliogenesis. (C) Confirmation by qPCR of the foxj1a, rfx2 and polaris (target gene) RNA levels in the whole embryo, normalized to beta-actin. MO = morpholino, WISH = whole-mount in situ hybridization, DFC = dorsal forerunner cell, KV = Kupffer’s vesicle.
Discussion
We found that Gpr22 affects KV cilia length and structure during zebrafish development. Overexpression or knock down of gpr22 in the zebrafish embryo resulted in a downwards curvature of the tail, heart edema and LR patterning defects. These phenotypes are frequently observed in zebrafish with mutations in genes involved in ciliogenesis [22]. Remarkably, zebrafish ciliary mutants all show the same range of phenotypes, independently on which gene is mutated or which step of ciliogenesis is affected. For example, the fleer mutant has a mutation in a gene involved in the post-translational polyglutamylation of tubulin and consequently has defects in cilia structure [38]. The oval and ift88 (polaris) mutants have a defective intraflagellar transport system resulting in changes in cilia length and structure [39], [40]. In the shmalhans mutant, cilia motility is affected due to a destabilization of the dynein arms [41]. Nonetheless, they are all characterized by a bent tail, heart edema, hydrocephalus, kidney cysts, otolith defects and randomized LR patterning. Besides the curved tail, the heart edema and the LR patterning defects, embryos with deregulated gpr22 expression also displayed otolith defects, suggesting that Gpr22 affects ciliogenesis in other organs besides the KV (Fig. S6) [42].
Any defects in KV cilia length or structure will lead to a disturbance of the directional fluid flow and thus randomized LR patterning, which can easily be used as a read-out for changes in KV ciliogenesis [24], [30]. Indeed, the randomized LR patterning defect observed in both gpr22 morphants and embryos with an overexpression of gpr22, was the result of shortening or lengthening, respectively, of the KV cilia and changes in the ultrastructure of the cilia.
We noticed that a DFC/KV specific knock down of gpr22 results in a milder reduction of KV cilia length (4.4±0.13 µm instead of 5.1±0.11 µm; P = <0.01), compared to a knock down in the whole embryo (3.4±0.12 µm instead of 5.4±0.16 µm; P = <0.01). Although we attempted to solve this issue by increasing the injected dose, it still might be that the MO did not diffuse efficiently into the DFCs, as it often aggregates at the site of injection. Another possible explanation is that an amount of functional Gpr22 protein is already present at the mid-blastula stage, which can partially compensate for the MO-blocked translation of the gpr22 RNA.
In contrast to the zebrafish gpr22 morphants, Gpr22 null mice display no obvious defects. However, the function of Gpr22 in ciliogenesis has not been specifically investigated in this study [7]. Further, it has been previously reported that there can be differences between zebrafish and mice in the requirement of cilia proteins. For example, a knock down of septin 6 or lethal giant larvae 2 results in cilia-associated phenotypes in zebrafish, while the mouse knock outs lack any phenotype [43], [44]. Additionally, GPCRs form oligomeric complexes, making it difficult to predict the outcome of a specific GPCR knock down and raising the possibility that GPCRs can function redundantly [45], [46].
Many of the characterized pathways such as FGF, Wnt or Notch, regulate KV cilia length in zebrafish by regulating the master regulators of ciliogenesis, Foxj1 or Rfx2 [47], [48]. Our data indicate, however, that Gpr22 does not alter the expression or transcriptional activity of neither of these transcription factors. It has been shown that overexpression of GPR22 in HEK-293 cells led to the inhibition of PKA activity [7] and that a decrease in PKA activity inhibits ciliogenesis and reduces cilia length [49]–[51]. However, embryos with an overexpression of gpr22, which should have a decrease in PKA activity, have the opposite phenotype. Thus, based on the literature, it is unlikely that Gpr22 signals in a PKA-depending manner to regulate KV cilia length.
Our finding that Gpr22 is involved in ciliogenesis, is supported by many studies that point towards a function of other GPCRs in ciliogenesis. GPCRs are often targeted to the cilia, such as the rhodopsin receptor in the outer segment of photoreceptors, the smoothened receptor controlling Hedgehog signaling in the primary cilia and the serotonin type 6 and somatostatin type 3 receptors in the neuronal cilia [12], [52], [53]. Further, it has been shown that the localization and trafficking of GPCRs in the neuronal cilia is regulated by the ciliary Bardet-Biedl syndrome proteins [54]. Additionally, conserved ciliary localization sequences have been found within GPCRs, like the AxxxQ motif in the IL3 and the VxPx motifs [12], [52], [55]. Finally, a chemical screen performed in Chlamydomonas identified the class A GPCRs as regulators of ciliogenesis [13]. The lack of a zebrafish Gpr22 antibody, however, prevented us from investigating the subcellular localization of the Gpr22 protein in zebrafish, but we were able to detect porcine Gpr22 in cilia protein extracts isolated from a porcine kidney epithelial cell line (CL4) (in collaboration with Wim Annaert and Applonia Rose, Laboratory for Membrane Transport, University of Leuven). Moreover, analysis of the Gpr22 protein sequence for cilia targeting motifs, revealed two VxPx and one AxxxQ motif, suggesting that Gpr22 might be targeted to the cilia. Nonetheless, many regulators of ciliogenesis are not targeted to the cilium per se, such as transcription factors, miRNAs, proteins associated with the centrosome, the basal bodies, actin etc. [25], [56]–[59].
Despite the growing list of genes and pathways affecting ciliogenesis, including Gpr22, the actual mechanism of maintaining cilia length and structure is still poorly investigated. Only theoretical models have been proposed, like the ‘balance-point’ mechanism [17]. In this model, the efficiency of the intraflagellar transport (IFT) is reversely proportional to the cilia length. Since the amount and distribution of IFT trains is the same in longer and shorter cilia, longer cilia will have smaller IFT trains that have to carry cargo along a longer distance. This results in the transport of less cargo to the tip and a net disassembly of the cilium. The steady-state length is reached when the assembly rate is the same as the disassembly rate. The question still remains how the amount of IFT particles is maintained in the cilium. Gpr22, together with genes already identified in screens for cilia length mutants, might control IFT quantity and therefore, cilia length and structure [17].
GPR22 is part of a gene cluster that has been genetically associated with human OA. Initial analysis revealed that GPR22 mRNA is present in both monolayer cultures (2D) and in pellets (3D) of human articular chondrocytes, indicating that GPR22 is also expressed in an environment that more accurately recapitulates articular cartilage. OA mouse models displayed expression of GPR22 in the articular cartilage of the knee joints [8], [9]. However, the function of GPR22 in the disease process is still controversial, since Raine et al. could not detect GPR22 RNA expression in human cartilage or joint tissues [8]–[10]. Nevertheless, our research could give more insight into the possible role of GPR22 in OA. Cilia are indeed necessary for proper cartilage/bone development and maintenance, mainly because of their essential role in Hedgehog, Wnt and mechano/chemosensing signaling pathways [60], [61]. It has been proposed that the primary cilia of chondrocytes act to maintain the columnar organization of the growth plate by a process termed chondrocyte rotation [62], [63]. The authors found that a conditional knock out of the IFT protein Kif3a in the chondrocytes of mice embryos, resulting in a lack of primary chondrocyte cilia, led to a disrupted columnar orientation in the growth plate and post-natal dwarfism [63]. Further, human ciliopathies are often characterized by skeletal defects, such as the Bardet-Biedl syndrome, the short rib-polydactyly group (SRPs) of ciliopathies and the Jeune syndrome (ATD) [61]. Moreover, Col2aCre;Ift88fl/fl mice, in which the primary cilia of the chondrocytes are lost, displayed symptoms of early OA because of a disturbed balance in the HH signaling pathway [64]. Additionally, it has been shown that the incidence and length of primary chondrocyte cilia increased at the degenerative articular surface in osteoarthritic tissue [65].
To conclude, our research identifies a novel player in the field of ciliogenesis and might open the door to new insights in the potential function of GPR22 in OA.
Materials and Methods
Zebrafish strains and maintenance
We confirm that all animal experiments were done according to the latest regulations set by the Belgian authorities that follow the European Parliament and Council Directive 2010/63/EU on the protection of animals used for scientific purposes. We confirm we also obtained approval by the Animal Care Committee of KU Leuven, the acting Institutional Animal Care and Use Committee (IACUC) of KU Leuven.
Adult zebrafish (Danio rerio) were maintained at 28.5°C, on a 14/10 hour, light/dark cycle under standard aquaculture conditions as described (www.zfin.org). All experiments were performed using embryos obtained from random matings of the wild type Oregon AB strain or the Tg(cmlc2:GFP) strain [29]. Embryos were staged by hours post-fertilization (hpf) and the number of somites according to Kimmel et al. [66].
Morpholino (MO) microinjections
Fluorescein tagged antisense MO oligonucleotides targeting the 5′UTR of gpr22 RNA and a control MO were obtained from Gene Tools, LLC (MO1: 5′-TCCCCTCCCTTGTGTTCCCTGTCCT-3′; MO2: 5′-ACTCCATCCATACTCCAATGCCTCT-3′; control MO: 5′-GGATTAAAATCCGCTACTCACATCC-3′; p53 MO: 5′-GCGCCATTGCTTTGCAAGAATTG-3′). Unless specified otherwise in the Results, 0.2 mM MO1 together with 0.1 mM p53 MO, or 0.4 mM MO2 were injected into the yolk of one cell stage embryos in a volume of 1 nl. To target MOs specifically to the dorsal forerunner cells (DFCs), 1 mM of MO was injected into the yolk at mid-blastula stage as previously described [32]. After injection, embryos with fluorescent-MO positive DFCs were selected at the 1–5 somite stage for further analysis.
Cloning and mRNA microinjections
The protein conservation between zebrafish and human GPR22 was determined by the program “specialized blast”, “align” (www.ncbi.nlm.nih.gov/BLAST/). We identified the 7TM domains using the hidden Markov model (TMHMM Server v. 2.0, http://www.cbs.dtu.dk/services/TMHMM/) [67]. Zebrafish cDNA was generated using the RevertAid First Strand cDNA Synthesis Kit (Fermentas) from total RNA extracted from 24-hpf wild-type embryos by the High Pure RNA Tissue Kit (Roche). Next, we used the generated cDNA as a template to amplify the full-length open reading frame of zebrafish gpr22. The resulting fragments were then cloned into the mammalian expression vector pCS2+ and sent out for sequence analysis to confirm the identity. Capped mRNA was synthesized from the pCS2+ plasmid using the mMESSAGE mMACHINE SP6 Kit (Ambion). Unless specified otherwise in the Results, overexpression studies were performed by injecting 75 pg/nl of gpr22 mRNA into the yolk of one cell stage embryos in a volume of 1 nl.
Whole mount in situ hybridization (WISH)
WISH was performed as described previously [68]. RNA antisense or sense probes were generated from linearized plasmids using SP6, T3 or T7 polymerase and a digoxigenin labeling mix (Roche).
Whole mount immunostaining
Embryos were fixed in 4% PFA or Dent’s fixative (80% methanol: 20% dimethyl sulfoxide) and kept ON at 4°C. Subsequently, we followed the protocol described previously for immunostaining [69]. Cilia were stained using monoclonal anti-acetylated tubulin antibody (Sigma) in a 1/1000 dilution. Alexa Fluor 555 Goat Anti-mouse (Life Technologies) was used as a secondary antibody in a 1/1000 dilution. Pictures from the KV cilia were obtained with a Leica DMR fluorescence microscope (Leica Microsystems) after flatmounting the embryos on glass slides in 90% glycerol and cilia length was measured by ImageJ software. Differences in average cilia length between conditions were evaluated using the two-tailed unpaired Student’s t-test. Results were considered to be significant when P<0.01.
Transmission electron microscopy (TEM)
Embryos were fixed in 0.1 M sodium-cacodylate buffer containing 2.5% glutaraldehyde and 2% PFA ON at 4°C. They were additionally fixed in 1% osmium tetroxide for 2 h at 4°C. After dehydration by ethanol series, we performed an ‘en bloc’ staining in 4% uranyl acetate for 30 min at 4°C. Subsequently, embryos were treated with propylene oxide for 2×15 min and embedded in 100% epoxy resin for 2 d at 60°C. 70 nm sections of the KV were post stained with 4% uranyl acetate and lead citrate before analysis with a JEM-2100 JEOL microscope.
qPCR
Total RNA was isolated and pooled from 40 deyolked bud stage embryos using the High Pure RNA Isolation Kit (Roche) and cDNA was synthesized according to the manufacturer’s protocol (Primescript, Takara). qPCR was performed with gene specific intron spanning primers using the SYBR Premix Ex Taq I kit (Takara). RNA levels were normalized to the housekeeping gene beta-actin.
Supporting Information
gpr22 knock down results in a dose-dependent curvature of the tail. (A–C) Lateral views with anterior to the left of zebrafish embryos at 24 hpf, injected at one cell stage with (A) water, (B) 0.2 mM gpr22 MO1 with 0.1 mM p53 MO or (C) without p53 MO. (D) Quantification of the tail phenotypes. (A, B, D) Knock down of gpr22 with either MO1 or MO2, results in a dose-dependent WT, mild, medium or severe downwards curvature of the tail. (B, C) Co-injection with p53 MO rescues the head necrosis caused by MO1. MO = morpholino, hpf = hours post-fertilization, n = number of analyzed embryos, WT = wild type.
(TIF)
gpr22 overexpression results in a dose-dependent curvature of the tail. (A–C) Lateral views with anterior to the left of zebrafish embryos at 24 hpf, injected at one cell stage with (A) water or (B) 75 pg/nl gpr22 RNA. (C) Quantification of the tail phenotypes. (A–C) Overexpression of gpr22 results in a dose-dependent WT, mild, medium or severe curvature of the tail. hpf = hours post-fertilization, n = number of analyzed embryos, WT = wild type.
(TIF)
gpr22 knock down results in randomized expression of the left-specific LPM markers. (A, B) Dorsal views with anterior to the top of embryos at the 20 somite stage. (A) WISH for lefty1 or (B) pitx2a. The expression of the LR markers lefty1 and pitx2a is randomized in gpr22 morphants (straight line). LPM = lateral plate mesoderm, WISH = whole mount in situ hybridization.MO = morpholino.
(TIF)
gpr22 overexpression results in defective LR patterning. (A) Ventral or (B, C) dorsal views with anterior to the top of embryos at (A, B) 4 dpf or (C) 20 somite stage. (A) Transgenic embryos expressing GFP under control of the cmlc2 promoter. gpr22 overexpression results in (from left to right) normal, absent, reversed or bicardial cardiac looping (not shown). (B) WISH for foxa3. The liver of water injected control embryos develops at the left side of the embryo. In contrast, the position of the liver in embryos injected with gpr22 RNA is randomized (straight line). (C) WISH for southpaw (spaw). The expression of the early LR marker southpaw is randomized in gpr22 injected embryos (straight line). V = ventricle, A = atrium, dpf = days post-fertilization, GFP = green fluorescence protein, cmlc2 = cardiac myosin light chain type 2, LR = left-right, WISH = whole mount in situ hybridization.
(TIF)
Expression analysis of gpr22 in zebrafish development. (A–D’) WISH for gpr22. (A, B, C, D) Lateral view with anterior to the left. (B’) Dorsal view of the tail bud region. (C’) Anterior view. (D’) Dorsal view with anterior to the top. (A, B) From shield to bud stage, gpr22 is ubiquitously expressed. (B’) From bud stage onwards, the expression pattern becomes more and more restricted to the axial structures and the developing KV (arrow). (C–D’) At later stages, gpr22 is expressed in several brain regions, the heart and some ciliated sensory organs, like (D) the retina and (D, D’) the nasal sac. WISH = whole-mount in situ hybridization, KV = Kupffer’s vesicle, hpf = hours post-fertilization, r = retina, ns = nasal sac.
(TIF)
gpr22 morphants display otolith defects. Left ear with anterior to the left of zebrafish embryos at 2 dpf, injected at one cell stage with (A) water or (B, C) 0.2 mM gpr22 MO. (A) Control embryos have 2 tethered otoliths (white asterisks) at the anterior and posterior poles of the otic vesicle. In contrast, gpr22 morphants display (B) fused otoliths or (C) an increased number of otoliths, which are not correctly positioned. dpf = days post-fertilization, MO = morpholino.
(TIF)
Acknowledgments
We are grateful to Camila Esguerra (Laboratory for Pharmaceutical Biology, Univ. of Leuven, Belgium), Jeroen Bakkers and Stefan Schulte-Merker (Hubrecht Insititute, Univ. Medical Center Utrecht, Netherlands), Nathalie Jurisch-Yaksi, Applonia Josephine Rose and Wim Annaert (Laboratory for Membrane Transport, Univ. of Leuven, Belgium), Jeffrey Amack (Department of Cell and Developmental Biology, Univ. of NY, USA), Joseph Yost (Department of Neurobiology and Anatomy, Univ. of Utah, USA), Xueying Lin (Department of Biochemistry and Molecular Biology, Mayo Clinic College of Medicine, Rochester, USA), Chiu-Ju Huang (Institute of Molecular and Cell Biology, National Taiwan University) for advice and sharing plasmids/antibody/zebrafish strains and Pieter Baatsen (CME, vzw Center for Biology and Disease, Univ. of Leuven) for the help with TEM.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the “TREAT-OA”- (Translational Research in Europe Applied Technologies for OsteoArthritis) consortium, funded by the Framework 7 European Research Program (FP7) (http://www.treatoa.eu/) and a IWT (Agency for Innovation by Science and 21 Technology) Strategic Basic Research grant (http://www.iwt.be/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang X, Eggert US (2013) Non-traditional roles of G protein-coupled receptors in basic cell biology. Molecular bioSystems 9: 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tang XL, Wang Y, Li DL, Luo J, Liu MY (2012) Orphan G protein-coupled receptors (GPCRs): biological functions and potential drug targets. Acta pharmacologica Sinica 33: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Defea K (2008) Beta-arrestins and heterotrimeric G-proteins: collaborators and competitors in signal transduction. British journal of pharmacology 153 Suppl 1S298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walther C, Ferguson SS (2013) Arrestins: role in the desensitization, sequestration, and vesicular trafficking of G protein-coupled receptors. Progress in molecular biology and translational science 118: 93–113. [DOI] [PubMed] [Google Scholar]
- 5. Hupfeld CJ, Olefsky JM (2007) Regulation of receptor tyrosine kinase signaling by GRKs and beta-arrestins. Annual review of physiology 69: 561–577. [DOI] [PubMed] [Google Scholar]
- 6. O'Dowd BF, Nguyen T, Jung BP, Marchese A, Cheng R, et al. (1997) Cloning and chromosomal mapping of four putative novel human G-protein-coupled receptor genes. Gene 187: 75–81. [DOI] [PubMed] [Google Scholar]
- 7. Adams JW, Wang J, Davis JR, Liaw C, Gaidarov I, et al. (2008) Myocardial expression, signaling, and function of GPR22: a protective role for an orphan G protein-coupled receptor. American journal of physiology Heart and circulatory physiology 295: H509–521. [DOI] [PubMed] [Google Scholar]
- 8. Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, et al. (2010) A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum 62: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, et al. (2011) Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis 70: 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raine EV, Wreglesworth N, Dodd AW, Reynard LN, Loughlin J (2012) Gene expression analysis reveals HBP1 as a key target for the osteoarthritis susceptibility locus that maps to chromosome 7q22. Ann Rheum Dis 71: 2020–2027. [DOI] [PubMed] [Google Scholar]
- 11. Malbon CC (2005) G proteins in development. Nature reviews Molecular cell biology 6: 689–701. [DOI] [PubMed] [Google Scholar]
- 12. Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K (2008) Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Molecular biology of the cell 19: 1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Avasthi P, Marley A, Lin H, Gregori-Puigjane E, Shoichet BK, et al. (2012) A chemical screen identifies class a g-protein coupled receptors as regulators of cilia. ACS chemical biology 7: 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun X, Haley J, Bulgakov OV, Cai X, McGinnis J, et al. (2012) Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia 1: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Molla-Herman A, Boularan C, Ghossoub R, Scott MG, Burtey A, et al. (2008) Targeting of beta-arrestin2 to the centrosome and primary cilium: role in cell proliferation control. PloS one 3: e3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrante MI, Romio L, Castro S, Collins JE, Goulding DA, et al. (2009) Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Human molecular genetics 18: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell's antenna. Nature reviews Molecular cell biology 12: 222–234. [DOI] [PubMed] [Google Scholar]
- 18. Avasthi P, Marshall WF (2012) Stages of ciliogenesis and regulation of ciliary length. Differentiation; research in biological diversity 83: S30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nature reviews Molecular cell biology 8: 880–893. [DOI] [PubMed] [Google Scholar]
- 20. Drummond I (2009) Studying cilia in zebrafish. Methods in cell biology 93: 197–217. [DOI] [PubMed] [Google Scholar]
- 21. Jaffe KM, Thiberge SY, Bisher ME, Burdine RD (2010) Imaging cilia in zebrafish. Methods in cell biology 97: 415–435. [DOI] [PubMed] [Google Scholar]
- 22. Malicki J, Avanesov A, Li J, Yuan S, Sun Z (2011) Analysis of cilia structure and function in zebrafish. Methods in cell biology 101: 39–74. [DOI] [PubMed] [Google Scholar]
- 23. Oteiza P, Koppen M, Concha ML, Heisenberg CP (2008) Origin and shaping of the laterality organ in zebrafish. Development 135: 2807–2813. [DOI] [PubMed] [Google Scholar]
- 24. Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ (2005) Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132: 1247–1260. [DOI] [PubMed] [Google Scholar]
- 25. Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, et al. (2010) Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biology of the cell/under the auspices of the European Cell Biology Organization 102: 499–513. [DOI] [PubMed] [Google Scholar]
- 26. Rovati GE, Capra V, Neubig RR (2007) The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Molecular pharmacology 71: 959–964. [DOI] [PubMed] [Google Scholar]
- 27. Konvicka K, Guarnieri F, Ballesteros JA, Weinstein H (1998) A proposed structure for transmembrane segment 7 of G protein-coupled receptors incorporating an asn-Pro/Asp-Pro motif. Biophysical journal 75: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, et al. (2007) p53 activation by knockdown technologies. PLoS genetics 3: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ (2003) Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn 228: 30–40. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Yost HJ, Amack JD (2013) Analysis of gene function and visualization of cilia-generated fluid flow in Kupffer's vesicle. Journal of visualized experiments: JoVE. [DOI] [PMC free article] [PubMed]
- 31. Thisse B, Heyer V, Lux A, Alunni V, Degrave A, et al. (2004) Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods in cell biology 77: 505–519. [DOI] [PubMed] [Google Scholar]
- 32. Amack JD, Yost HJ (2004) The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol 14: 685–690. [DOI] [PubMed] [Google Scholar]
- 33. Matsui T, Thitamadee S, Murata T, Kakinuma H, Nabetani T, et al. (2011) Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer's vesicle organogenesis. Proc Natl Acad Sci U S A 108: 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexander J, Rothenberg M, Henry GL, Stainier DY (1999) casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol 215: 343–357. [DOI] [PubMed] [Google Scholar]
- 35. Hong SK, Dawid IB (2009) FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc Natl Acad Sci U S A 106: 2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hashimoto H, Rebagliati M, Ahmad N, Muraoka O, Kurokawa T, et al. (2004) The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 131: 1741–1753. [DOI] [PubMed] [Google Scholar]
- 37. Francescatto L, Rothschild SC, Myers AL, Tombes RM (2010) The activation of membrane targeted CaMK-II in the zebrafish Kupffer's vesicle is required for left-right asymmetry. Development 137: 2753–2762. [DOI] [PubMed] [Google Scholar]
- 38. Pathak N, Obara T, Mangos S, Liu Y, Drummond IA (2007) The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Molecular biology of the cell 18: 4353–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, et al. (2005) Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132: 1907–1921. [DOI] [PubMed] [Google Scholar]
- 40. Lunt SC, Haynes T, Perkins BD (2009) Zebrafish ift57, ift88, and ift172 intraflagellar transport mutants disrupt cilia but do not affect hedgehog signaling. Dev Dyn 238: 1744–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Panizzi JR, Becker-Heck A, Castleman VH, Al-Mutairi DA, Liu Y, et al. (2012) CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet 44: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stooke-Vaughan GA, Huang P, Hammond KL, Schier AF, Whitfield TT (2012) The role of hair cells, cilia and ciliary motility in otolith formation in the zebrafish otic vesicle. Development 139: 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhai G, Gu Q, He J, Lou Q, Chen X, et al. (2014) Sept6 is required for ciliogenesis in Kupffer's vesicle, the pronephros, and the neural tube during early embryonic development. Molecular and cellular biology 34: 1310–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tay HG, Schulze SK, Compagnon J, Foley FC, Heisenberg CP, et al. (2013) Lethal giant larvae 2 regulates development of the ciliated organ Kupffer's vesicle. Development 140: 1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rivero-Muller A, Jonas KC, Hanyaloglu AC, Huhtaniemi I (2013) Di/oligomerization of GPCRs-mechanisms and functional significance. Progress in molecular biology and translational science 117: 163–185. [DOI] [PubMed] [Google Scholar]
- 46. Van Craenenbroeck K (2012) GPCR oligomerization: contribution to receptor biogenesis. Sub-cellular biochemistry 63: 43–65. [DOI] [PubMed] [Google Scholar]
- 47. Caron A, Xu X, Lin X (2012) Wnt/beta-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer's vesicle. Development 139: 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lopes SS, Lourenco R, Pacheco L, Moreno N, Kreiling J, et al. (2010) Notch signalling regulates left-right asymmetry through ciliary length control. Development 137: 3625–3632. [DOI] [PubMed] [Google Scholar]
- 49. Abdul-Majeed S, Moloney BC, Nauli SM (2012) Mechanisms regulating cilia growth and cilia function in endothelial cells. Cellular and molecular life sciences: CMLS 69: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wann AK, Knight MM (2012) Primary cilia elongation in response to interleukin-1 mediates the inflammatory response. Cellular and molecular life sciences: CMLS 69: 2967–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Besschetnova TY, Kolpakova-Hart E, Guan Y, Zhou J, Olsen BR, et al. (2010) Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr Biol 20: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lodowski KH, Lee R, Ropelewski P, Nemet I, Tian G, et al. (2013) Signals governing the trafficking and mistrafficking of a ciliary GPCR, rhodopsin. The Journal of neuroscience: the official journal of the Society for Neuroscience 33: 13621–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, et al. (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437: 1018–1021. [DOI] [PubMed] [Google Scholar]
- 54. Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K (2008) Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A 105: 4242–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Geng L, Okuhara D, Yu Z, Tian X, Cai Y, et al. (2006) Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci 119: 1383–1395. [DOI] [PubMed] [Google Scholar]
- 56. Song R, Walentek P, Sponer N, Klimke A, Lee JS, et al. (2014) miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510: 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cao J, Shen Y, Zhu L, Xu Y, Zhou Y, et al. (2012) miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat Cell Biol 14: 697–706. [DOI] [PubMed] [Google Scholar]
- 58. Lee JY, Stearns T (2013) FOP is a centriolar satellite protein involved in ciliogenesis. PloS one 8: e58589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilkinson CJ, Carl M, Harris WA (2009) Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC cell biology 10: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Haycraft CJ, Serra R (2008) Cilia involvement in patterning and maintenance of the skeleton. Curr Top Dev Biol 85: 303–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huber C, Cormier-Daire V (2012) Ciliary disorder of the skeleton. American journal of medical genetics Part C, Seminars in medical genetics 160C: 165–174. [DOI] [PubMed] [Google Scholar]
- 62. Ascenzi MG, Blanco C, Drayer I, Kim H, Wilson R, et al. (2011) Effect of localization, length and orientation of chondrocytic primary cilium on murine growth plate organization. Journal of theoretical biology 285: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, et al. (2007) Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development 134: 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang CF, Ramaswamy G, Serra R (2012) Depletion of primary cilia in articular chondrocytes results in reduced Gli3 repressor to activator ratio, increased Hedgehog signaling, and symptoms of early osteoarthritis. Osteoarthritis Cartilage 20: 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McGlashan SR, Cluett EC, Jensen CG, Poole CA (2008) Primary cilia in osteoarthritic chondrocytes: from chondrons to clusters. Dev Dyn 237: 2013–2020. [DOI] [PubMed] [Google Scholar]
- 66. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- 67. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 68. Thisse C, Thisse B (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols 3: 59–69. [DOI] [PubMed] [Google Scholar]
- 69. Monteiro R, van Dinther M, Bakkers J, Wilkinson R, Patient R, et al. (2008) Two novel type II receptors mediate BMP signalling and are required to establish left-right asymmetry in zebrafish. Dev Biol 315: 55–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gpr22 knock down results in a dose-dependent curvature of the tail. (A–C) Lateral views with anterior to the left of zebrafish embryos at 24 hpf, injected at one cell stage with (A) water, (B) 0.2 mM gpr22 MO1 with 0.1 mM p53 MO or (C) without p53 MO. (D) Quantification of the tail phenotypes. (A, B, D) Knock down of gpr22 with either MO1 or MO2, results in a dose-dependent WT, mild, medium or severe downwards curvature of the tail. (B, C) Co-injection with p53 MO rescues the head necrosis caused by MO1. MO = morpholino, hpf = hours post-fertilization, n = number of analyzed embryos, WT = wild type.
(TIF)
gpr22 overexpression results in a dose-dependent curvature of the tail. (A–C) Lateral views with anterior to the left of zebrafish embryos at 24 hpf, injected at one cell stage with (A) water or (B) 75 pg/nl gpr22 RNA. (C) Quantification of the tail phenotypes. (A–C) Overexpression of gpr22 results in a dose-dependent WT, mild, medium or severe curvature of the tail. hpf = hours post-fertilization, n = number of analyzed embryos, WT = wild type.
(TIF)
gpr22 knock down results in randomized expression of the left-specific LPM markers. (A, B) Dorsal views with anterior to the top of embryos at the 20 somite stage. (A) WISH for lefty1 or (B) pitx2a. The expression of the LR markers lefty1 and pitx2a is randomized in gpr22 morphants (straight line). LPM = lateral plate mesoderm, WISH = whole mount in situ hybridization.MO = morpholino.
(TIF)
gpr22 overexpression results in defective LR patterning. (A) Ventral or (B, C) dorsal views with anterior to the top of embryos at (A, B) 4 dpf or (C) 20 somite stage. (A) Transgenic embryos expressing GFP under control of the cmlc2 promoter. gpr22 overexpression results in (from left to right) normal, absent, reversed or bicardial cardiac looping (not shown). (B) WISH for foxa3. The liver of water injected control embryos develops at the left side of the embryo. In contrast, the position of the liver in embryos injected with gpr22 RNA is randomized (straight line). (C) WISH for southpaw (spaw). The expression of the early LR marker southpaw is randomized in gpr22 injected embryos (straight line). V = ventricle, A = atrium, dpf = days post-fertilization, GFP = green fluorescence protein, cmlc2 = cardiac myosin light chain type 2, LR = left-right, WISH = whole mount in situ hybridization.
(TIF)
Expression analysis of gpr22 in zebrafish development. (A–D’) WISH for gpr22. (A, B, C, D) Lateral view with anterior to the left. (B’) Dorsal view of the tail bud region. (C’) Anterior view. (D’) Dorsal view with anterior to the top. (A, B) From shield to bud stage, gpr22 is ubiquitously expressed. (B’) From bud stage onwards, the expression pattern becomes more and more restricted to the axial structures and the developing KV (arrow). (C–D’) At later stages, gpr22 is expressed in several brain regions, the heart and some ciliated sensory organs, like (D) the retina and (D, D’) the nasal sac. WISH = whole-mount in situ hybridization, KV = Kupffer’s vesicle, hpf = hours post-fertilization, r = retina, ns = nasal sac.
(TIF)
gpr22 morphants display otolith defects. Left ear with anterior to the left of zebrafish embryos at 2 dpf, injected at one cell stage with (A) water or (B, C) 0.2 mM gpr22 MO. (A) Control embryos have 2 tethered otoliths (white asterisks) at the anterior and posterior poles of the otic vesicle. In contrast, gpr22 morphants display (B) fused otoliths or (C) an increased number of otoliths, which are not correctly positioned. dpf = days post-fertilization, MO = morpholino.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.