Abstract
The human leukocyte antigen (HLA) class I and class II loci are the most polymorphic genes in the human genome. Hematopoietic stem cell transplantation requires allele-level HLA typing at multiple loci to select the best matched unrelated donors for recipient patients. In current methods for HLA typing, both alleles of a heterozygote are amplified and typed or sequenced simultaneously, often making it difficult to unambiguously determine the sequence of the two alleles. Next-generation sequencing methods clonally propagate in parallel millions of single DNA molecules, which are then also sequenced in parallel. Recently, the read lengths obtainable by one such next-generation sequencing method (454 Life Sciences, Inc.) have increased to >250 nucleotides. These clonal read lengths make possible setting the phase of the linked polymorphisms within an exon and thus the unambiguous determination of the sequence of each HLA allele. Here we demonstrate this capacity as well as show that the throughput of the system is sufficiently high to enable a complete, 7-locus HLA class I and II typing for 24 or 48 individual DNAs in a single GS FLX sequencing run. Highly multiplexed amplicon sequencing is facilitated by the use of sample-specific internal sequence tags (multiplex identification tags or MIDs) in the primers that allow pooling of samples yet maintain the ability to assign sequences to specific individuals. We have incorporated an HLA typing software application developed by Conexio Genomics (Freemantle, Australia) that assigns HLA genotypes for these 7 loci (HLA-A, -B, -C, DRB1, DQA1, DQB1, DPB1), as well as for DRB3, DRB4, and DRB5 from 454 sequence data. The potential of this HLA sequencing system to analyze chimeric mixtures is demonstrated here by the detection of a rare HLA-B allele in a mixture of two homozygous cell lines (1/100), as well as by the detection of the rare nontransmitted maternal allele present in the blood of a severe combined immunodeficiency disease syndrome (SCIDS) patient.
Keywords: 454, human leukocyte antigen, sequencing
Introduction
The human leukocyte antigen (HLA) class I and class II loci are the most polymorphic genes in the human genome, with a complex pattern of patchwork polymorphism localized primarily to exon 2 for the class II genes and exons 2 and 3 for the class I genes. For current HLA typing methods, allele-level resolution of HLA alleles, which is clinically important for hematopoetic stem cell (HSC) transplantation in the unrelated donor setting, is technically challenging (see below). Several large-scale studies have demonstrated that precise, allele-level HLA matching between donor and patient significantly improves overall transplant survival by reducing the incidence and severity of both acute and chronic GVHD (graft versus host disease) and improving the rates of successful engraftment (1–15).
Currently, bone marrow donor registries contain data on millions of potential donors who have been analyzed, for the most part, at an intermediate level of resolution for HLA- A, -B, and DRB1 loci. Multiple potentially matched unrelated donors are selected, based on this initial typing, and then these donor samples are reanalyzed at allele-level resolution at these and additional HLA loci to identify the donor best matched to the recipient.
Currently, the highest resolution HLA typing is obtained with fluorescent, Sanger-based DNA sequencing using capillary electrophoresis. Ambiguities in the HLA typing data may still persist due to multiple polymorphisms shared between alleles and the resultant phase ambiguities when both alleles are amplified and sequenced together. Resolving these ambiguities requires time-consuming approaches such as amplifying and then analyzing the two alleles separately.
An alternative approach to the phase ambiguity problem is clonal sequencing. Next-generation DNA sequencing (16) provides orders of magnitude increases in the number of reads of contiguous sequence obtainable in a short time. Sequencing hundreds of millions of bases from amplified single DNA molecules is possible within a few days. To date, however, read lengths that would allow the resolution of phase ambiguities in HLA alleles have been achieved only with the clonal pyrosequencing-based method developed by 454 Life Sciences, Inc (17). The Roche GS FLX genome sequencer generates sequence read lengths greater than 250 nucleotides. The average HLA exon encoding the peptide binding groove is approximately 270 base pairs; the range is 239–242bp for DQA1 exon 2 to 276bp for exon 3 of class I genes. As our amplicons are only slightly longer than the exons, each exon can be sequenced completely by sequencing both strands with sufficient overlap between the reads that specific HLA alleles can be unambiguously assigned. We have chosen to analyze HLA polymorphism by isolating the relevant exons through specific polymerase chain reaction (PCR) amplification, prior to emulsion PCR and pyrosequencing rather than capturing by hybridization and then sequencing the relevant genomic DNA.
Here, we demonstrate that this approach allows the rapid, accurate determination of HLA type at allelic resolution for many individuals at multiple HLA loci simultaneously. This approach features novel HLA genotyping software developed by Conexio Genomics, Inc. (Freemantle, Australia) for analyzing sequence read data from the Roche GS FLX instrument (specifically, the fna files). This software compares the sequence reads to the database of known HLA allele sequences and assigns a genotype for each locus for each individual.
The very large number of sequence reads (n = 300 – 400K) generated in a single run makes possible the detection of rare sequence variants present in individual samples. For example, maternal cells can be found in low frequencies in the blood of some severe combined immunodeficiency disease syndrome (SCIDS) patients; these chimeric mixtures, consequently, contain rare nontransmitted maternal alleles. Here, we demonstrate this capability of 454 sequencing through the analysis of DNA mixtures from two homozygous cell lines, as well as through the analysis of DNA from an SCIDS patient. In this case, rare copies of the maternal nontransmitted allele could be detected, in addition to the inherited paternal and maternal alleles at the HLA-B and HLA-C loci.
Materials and methods
Primer design and PCR conditions
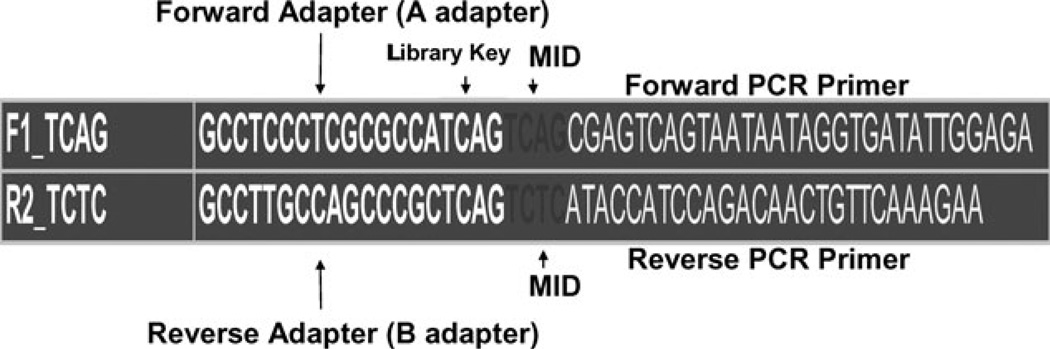
The 454 HLA fusion primers consist of four main parts (Figure 1). Starting from the 5’ end, the primer contains a 19-base adapter sequence, which is responsible for capture of PCR amplicons by DNA capture beads. Adapter sequences end with a 4-base library key tag (TCAG), which allows the 454-genome sequencer software to differentiate HLA amplicon derived sequences from internal control sequences. We added 4-base multiplex identifier (MID) sequences (18) immediately following the library key tag to allow for multiplexed sequencing of HLA amplicons. The locus-specific sequence for amplification of the target genomic region follows the MID sequence (see Table S1, Supporting Information) for the HLA locus-specific primer sequences). Fusion primers were designed in sets of 12, with each primer having a unique MID sequence. The design of these primers involves the usual ‘trade-offs’ for HLA amplification; the primers should be specific to the locus, to the extent possible, and also be capable of amplifying all alleles at that locus with comparable efficiency. If the 454 HLA fusion primers are not completely specific (for example, an HLA-A exon 4 primer pair could also amplify HLA-E, -F or -G), then, unlike the case with Sanger sequencing or SSOP typing methods where sequences of related genes adds ‘noise’ to the typing system, these sequence reads can be filtered out such that the genotype assignment is unaffected. In some cases, however, as in the coamplification of DRB3, DRB4, and DRB5 together with the DRB1 locus using generic DRB primers, these additional sequence reads can serve as potentially important genetic markers and provide additional valuable genotypes.
Figure 1.
Schematic of 454 sequencing fusion primer pair with 4-base multiplex identifier (MIDs).
The PCR amplifications of 14 exons from the 24 cell-line DNAs were all carried out individually. The thermal cycling conditions are as follows: 95°−10’, 95°−15”, 60°−45”, 72°−15”; 35 cycles, 72°−5’. We note that our HLA-C-specific exon 3 primers used in this experiment generate a 653-bp amplicon. This amplicon is too long to allow complete sequencing of exon 3 by the GS FLX (average read length is 250 bases). Using this amplicon as the template for nested PCR with primers FDB1180 and RHLACE3 (Table S1, Supporting Information) generates a 381-bp amplicon from which full coverage sequencing can be achieved. Currently, we use only the second ‘internal’ primer pair yielding a 381-bp amplicon directly from genomic DNA, so that a nested PCR is not necessary.
Each of the 336 PCR reactions (25 ul) was prepared using a standard master mix that consisted of 10 mM Tris-HCl buffer, pH 8.3, 50 mM KCl, 1.5 mM MgCl, 150 uM dNTPs, Glycerol 10% v/v, AmpliTaq Gold (2 units). This mix was then added to each well of a microtiter plate containing 10 ng of cell-line genomic DNA, and forward/reverse fusion primers (10 pmoles each). Following PCR amplification, nonspecific and primer–dimer artifact products were removed from the amplicons using the Agencourt AMPure system (Agencourt Bioscience Corp., Beverly, MA). Aliquots from purified amplicons are further evaluated by electrophoresis on eGel® s (Invitrogen Corporation, Carlsbad, CA). The amplicons are then quantified by Quant-iT™PicoGreen® assay (Invitrogen Corporation) on a Molecular Devices Gemini XS microplate fluorimeter (Molecular Devices, Sunnyvale, CA) and subsequently diluted to 200 000 molecules/µl. With 12 MIDs available per amplicon, the maximum number of samples that can be sequenced in a picotitre plate region is 12. Thus, in our 24-sample runs, we created two pools of amplicons for all loci, one pool for samples 1–12 and another for samples 13–24. Emulsion PCR, bead recovery, and pyrosequencing were carried out as described (Roche Applied Science GS FLX emPCR Method Manual USM-00033.A-December 2007, Roche Applied Science GS FLX Sequencing Method Manual USM-00035.A-December 2007).
Results
Multiplex pyrosequencing
The analysis of multiple HLA loci for multiple samples in a single 454 run is facilitated by the incorporation of MID tags into the PCR fusion primers (18). Figure 1 illustrates the 454 fusion primer structure, and Table S1 (Supporting Information) presents the sequences of the HLA-specific primers (without MID or adapter sequences) that were used to generate the data reported here. Fourteen primer pairs were designed for exons 2, 3, and 4 of HLA-A, B, and C, exon 2 of DRB1, DPB1, DQA1, and exons 2 and 3 of DQB1. Primers with 12 different MID tags for each target sequence were designed for a total of 168 (14 × 12) primer pairs. The primers for exon 2 of DRB1 also amplify the DRB3, DRB4, and DRB5 loci, genes that are present on specific DRB1 haplotypes (http://www.ebi.ac.uk/imgt/hla/).
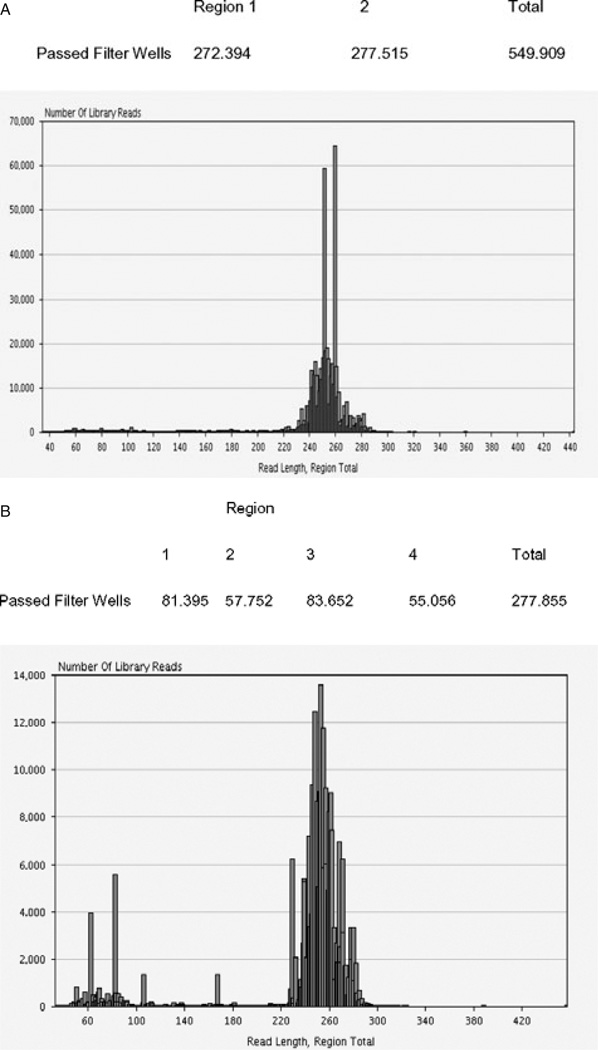
Following amplification of the various samples, the PCR products were quantified by PicoGreen fluorescence, diluted to the appropriate concentration, and pooled for emulsion PCR. Pyrosequencing runs of 24 and 48 individuals were achieved using 2 or 4 picotiter plate regions, respectively. Average read depths for each exon (per individual sample) are shown in Table 1. Overall average read depths per amplicon range from 500 to 700 reads (forward sequences + reverse sequences). Typical HLA amplicon passed filter sequence read yields and read length distributions for amplicons in a 24-sample run (336 amplicons) are shown in Figure 2(A) and (B) for a 48-sample run (672 amplicons). Read length distributions are centered around the 250 bases. This length is sufficient for forward and reverse sequence reads to overlap, allowing unambiguous assignment of sequences to each exon and, ultimately, to each allele. The alignment of HLA sequences to the database of known HLA alleles and assignment of HLA genotypes is accomplished with the Conexio Genomics HLA genotyping software. In the sequence read length distribution from the 48-sample run (Figure 2B), a proportion of reads are short, ranging in size from 50 to 180 bases. The most numerous of these are in the 60–80 base range. These short sequences are the consequence of primer–dimer artifact from the initial PCR reactions that was carried into emulsion PCR with the diluted amplicon pools. The Agencourt AMPure system (Agencourt Bioscience Corp., Beverly, MA) was not used to purify the primer–dimer artifact from the amplicons used in this particular experiment, while it was used to purify the amplicons sequenced in the 24-sample run. A comparison of the read distributions between these two runs reveals the efficiency of the Agencourt AMPure system to remove primer–dimer artifact and any other shorter nonspecific PCR products, from amplicons being prepared for 454 sequencing.
Table 1.
Average number of forward (F) and reverse (R) sequence reads per amplicon per individual sample (24-sample run)
| HLA-A E2 | HLA-A E3 | HLA-A E4 | |
| F | 277 | 205 | 114 |
| R | 256 | 182 | 108 |
| HLA-B E2 | HLA-B E3 | HLA-B E4 | |
| F | 447 | 705 | 767 |
| R | 444 | 647 | 674 |
| HLA-C E2 | HLA-C E3 | HLA-C E4 | |
| F | 234 | 161 | 345 |
| R | 226 | 146 | 231 |
| DQA1 | DQB1 E2 | DQB1 E3 | |
| F | 634 | 595 | 576 |
| R | 575 | 506 | 576 |
| DPB1 | DRB1 | ||
| F | 290 | 206 | |
| R | 276 | 201 |
Average read depths per amplicon (24-sample run).
Figure 2.
(A) Typical HLA amplicon passed filter wells (sequence reads) by picotiter plate region and by read length (336 amplicons). (B) Typical HLA amplicon passed filterwells (sequence reads) by picotiter plate region and by read length (672 amplicons) for 48-sample run.
Genotyping software
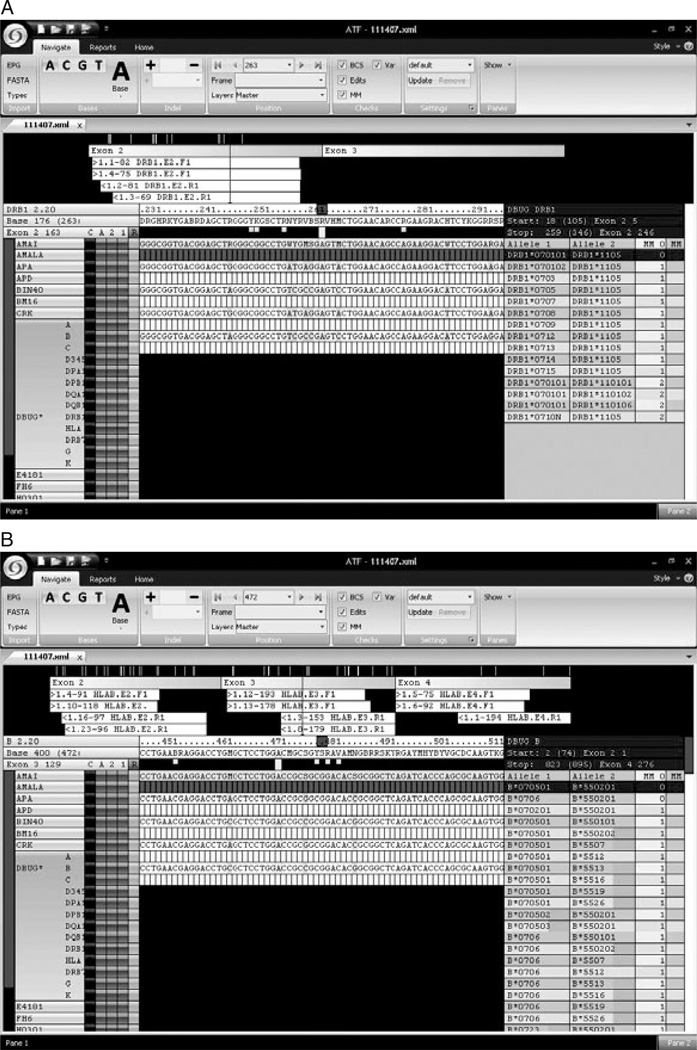
The GS FLX data processing software filters the hundreds of thousands of individual sequence reads generated in each sequencing run based on sequence quality length minimums resulting in sets of sequence reads that constitute the ‘passed filter’ reads. To facilitate HLA genotype assignment from 454 sequence data files, Conexio Genomics’ HLA genotyping software application compares the passed filter forward and reverse 454 sequence reads derived from each exon to the current IMGT-HLA sequence database (EMBL-European Bioinformatics Institute, Cambridge, UK). The database also contains the sequence of HLA pseudogenes and related genes, allowing the filtering out of sequences generated from pseudogenes or from nonclassical HLA class I genes (e.g. HLA-,E,F,G, and H). Screenshots of the software displaying the analysis of sequence reads for exon 2 of the DRB1 gene, and exons 2, 3, and 4 of the HLA-B genes for the cell-line DBUG are shown in Figures 3(A) and (B). The number of different forward and reverse sequence reads for each exon is shown in the upper panel. For the DRB1 exon 2, there were 82 forward reads of one allele (designated as sequence 1.1) and 75 forward reads of the other allele (designated as sequence 1.4) and 81 reverse reads of one allele (designated as 1.2) and 69 reads of the other allele (designated as 1.3). The genotype assignment is shown to the right, along with the number of mismatches of the sequence file to the HLA alleles in the database. In some cases, a unique genotype (top line in the right panel; DRB1*070101/1105) is assigned with 0 mismatches, as in Figure 3(A) for DRB1; other closely related potential genotype assignments having one or more mismatches are shown just below the 0 mismatch genotype assignment. In other cases, more than one possible genotype is consistent with the sequence data. In the sequence reads for DBUG for HLA-B exons 2, 3 and 4, two genotypes are assigned with 0 mismatches (Figure 3B). In this case, the polymorphism that distinguishes the two genotype assignments (B*070501 and B*070601) is located in exon 5 (not sequenced in this panel).
Figure 3.
(A) Screenshot of Conexio genotype assignment: DRB1 for DBUG. (B) Screenshot of Conexio genotype assignment: HLA-B for DBUG.
Rare sequence reads, such as those derived from pseudogenes, related HLA genes, or from PCR amplification or pyrosequencing artifacts, that differ from consensus allele sequences, are filtered into a ‘secondary alignment’. Analysis of these low-frequency sequences in the secondary alignment can prove instructive with regard to PCR primer specificity and systematic pyrosequencing errors. The most common, albeit still very rare, sequencing artifact we observed was variation in homopolymeric runs of G. For example, in the analysis of DNA from the AMALA cell line, we observed 283 reverse exon 2 HLA-A sequence reads for the consensus sequence of 4 Gs, while in the secondary alignment, we observed 9 reverse exon 2 reads for 3 Gs. Since the Conexio genotyping software filters these sequence reads into the secondary alignment, these rare sequence artifacts do not affect the accuracy and reliability of the HLA genotype assignments.
High-throughput HLA sequencing
A total of 24 cell-line derived DNA samples of known HLA type, based on previous analyses of probe hybridization HLA typing and Sanger sequencing results, were sequenced at all 7 loci (HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, DPB1). Exon 2 sequences of DRB3, DRB4, and DRB5 were also identified in the amplicons generated by the generic DRB primer pair. Subsequently, a run of 48 samples (24 cell-line DNAs and 24 DNAs extracted from blood samples) were sequenced at the same loci and genotype assignments were generated from the sequence data by the Conexio Genomics HLA genotyping software. In general, consensus sequences derived from 50 or more sequence reads are considered ‘high confidence’ as are the genotype assignments based on these consensus sequences. In the 24-sample run, the concordance of software genotype calls and previously determined HLA types for all 7 loci was 99.4% (Table 2). The allele assignments for DRB3, DRB4, and DRB5, are not included in this Table. If more than one genotype was assigned (ambiguity string) for a given locus, only the top genotype in the list (see Figure 4) is shown in the Table. Of the 168 allele calls (7 loci×24 samples), one was incorrect. The HLA-A*2601 allele of cellline DBUG (A*1101,*2601) was not called in this experiment by the software. DNAs from 24 cell lines were amplified by 454 fusion primers containing MID tags and analyzed in two regions as described in the Materials and Methods. The genotype assignments for these cell lines, based on SSOP or Sanger SBT (sequence-based typing) are shown in the two left panels. The genotype assignments determined by 454 sequencing and Conexio software are shown on the right. If the software indicated more than one genotype was consistent with the sequence data (ambiguity) and the correct reference genotype was included, we called this typing ‘concordant’. Concordance was 99.4% for the 168 allele calls (7 loci × 24 samples). For the cell-line DBUG (A*1101,*2601), the software correctly called the A*1101 allele but not the A*2601 allele in this run. Of the 167 correct allele calls, 133 were called correctly by the software with no manual editing, 26 were called correctly after the manual editing, and 9 were called correctly following nontarget gene (pseudogene, related HLA gene) removal.
Table 2.
Concordance of 454 GS FLX determined human leukocyte antigen (HLA) genotypes with reference genotypes
| Cell line | Locus | Allele1 | Allele2 | Allele1 454 |
Allele2 454 |
Cell line |
Locus | Allele1 | Allele2 | Allele1 454 |
Allele2 454 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| JW5 | DRB1 | 0103 | 03 | 0103 | 030101 | E4181324 | DRB1 | 150201 | 150201 | 150201 | 150201 |
| JW5 | DQA1 | 0101 | 0501 | 010101 | 050101 | E4181324 | DQA1 | 0103 | 0103 | 0103 | 0103 |
| JW5 | DQB1 | 0201/2 | 0501 | 020101 | 050101 | E4181324 | DQB1 | 060101 | 060101 | 060101 | 060101 |
| JW5 | DPB1 | 010101 | 020102 | 010101 | 020102 | E4181324 | DPB1 | 020102 | 0401 | 020102 | 040101 |
| JW5 | HLA-A | 0101 | 2301 | 01010101 | 2301 | E4181324 | HLA-A | 0101 | 0101 | 01010101 | 01010101 |
| JW5 | HLA-B | 0801/4 | 18 | 080101 | 180101 | E4181324 | HLA-B | 520101 | 520101 | 520101 | 520101 |
| JW5 | HLA-C | 05 | 07 | 050101 | 070101 | E4181324 | HLA-C | 1202 | 1202 | 120201 | 120202 |
| RAJI | DRB1 | 0301 | 100101 | 030101 | 100101 | SAVC | DRB1 | 0401 | 0401 | 040101 | 040101 |
| RAJI | DQA1 | 0101 | 0501 | 010101 | 0501 | SAVC | DQA1 | 0301 | 0301 | 030101 | 030101 |
| RAJI | DQB1 | 0201/2 | 0501 | 020101 | 050101 | SAVC | DQB1 | 0302 | 0302 | 030201 | 030201 |
| RAJI | DPB1 | 010101 | 010101 | 010101 | 010101 | SAVC | DPB1 | 1001 | 1001 | 1001 | 1001 |
| RAJI | HLA-A | 03 | 03 | 03010101 | 03010101 | SAVC | HLA-A | 0301 | 0301 | 03010101 | 03010101 |
| RAJI | HLA-B | 1510 | 1510 | 1510 | 1510 | SAVC | HLA-B | 0702 | 0702 | 070201 | 070201 |
| RAJI | HLA-C | 030402 | 04 | 030402 | 04010101 | SAVC | HLA-C | 0702 | 0702 | 07020101 | 07020101 |
| NAMALWA | DRB1 | 0405 | 1503 | 040501 | 1503 | LADA | DRB1 | 090102 | 1201/6 | 090102 | 120101 |
| NAMALWA | DQA1 | 0102 | 0301 | 010201 | 030101 | LADA | DQA1 | 0101 | 0301 | 010101 | 030101 |
| NAMALWA | DQB1 | 0302 | 0602 | 030201 | 0602 | LADA | DQB1 | 0201/2 | 0501 | 0202 | 050101 |
| NAMALWA | DPB1 | 0101 | 0201 | 010101 | 020102 | LADA | DPB1 | 0301 | 1701 | 030101 | 1701 |
| NAMALWA | HLA-A | 03 | 6802 | 03010101 | 68020101 | LADA | HLA-A | 0201 | 8001 | 02010101 | 8001 |
| NAMALWA | HLA-B | 0702 | 4901 | 070201 | 4901 | LADA | HLA-B | 0702 | 5703 | 070201 | 570301 |
| NAMALWA | HLA-C | 0701/6 | 0702/3 | 070101 | 07020101 | LADA | HLA-C | 0702/3 | 0802 | 07020101 | 0802 |
| APA | DRB1 | 1405 | 150101/102 | 140501 | 150101 | DBUG | DRB1 | 0701 | 1105 | 070101 | 1105 |
| APA | DQA1 | 0101 | 0102 | 010101 | 010201 | DBUG | DQA1 | 0101 | 0201 | 010101 | 0201 |
| APA | DQB1 | 050301 | 0601 | 050301 | 060101 | DBUG | DQB1 | 030302 | 0602 | 030302 | 0602 |
| APA | DPB1 | 0501 | 0501 | 0501 | 0501 | DBUG | DPB1 | 040101 | 0501 | 040101 | 0501 |
| APA | HLA-A | 2403 | 1101 | 110101 | 240301 | DBUG | HLA-A | 1101 | 2601 | 1101 | |
| APA | HLA-B | 1502 | 5502 | 1502 | 550201 | DBUG | HLA-B | 0705/6 | 55 | 070501 | 550201 |
| APA | HLA-C | 08 | 1203/6 | 080101 | 12030101 | DBUG | HLA-C | 010202 | 07020102 | 010201 | 07020101 |
| MG | DRB1 | 0401/16 | 1001 | 040101 | 100101 | AMAI | DRB1 | 1503 | 1503 | 1503 | 1503 |
| MG | DQA1 | 0101 | 0301 | 010201 | 030101 | AMAI | DQA1 | 0102 | 0102 | 010201 | 010201 |
| MG | DQB1 | 0302/7 | 0501 | 030201 | 050101 | AMAI | DQB1 | 0602 | 0602 | 0602 | 0602 |
| MG | DPB1 | 0401 | 0601 | 040101 | 0601 | AMAI | DPB1 | 0402 | 0402 | 0402 | 0402 |
| MG | HLA-A | 0101 | 0201 | 01010101 | 02010101 | AMAI | HLA-A | 6802 | 6802 | 68020101 | 68020101 |
| MG | HLA-B | 15 | 3701 | 15010101 | 370101 | AMAI | HLA-B | 5301 | 5301 | 530101 | 530101 |
| MG | HLA-C | 03 | 0602 | 030401 | 06020101 | AMAI | HLA-C | 0401 | 0401 | 040101 | 040101 |
| TTL | DRB1 | 1301 | 1501 | 130101 | 150101 | CRK | DRB1 | 0701 | 0701 | 070101 | 070101 |
| TTL | DQA1 | 0102 | 0103 | 010201 | 0103 | CRK | DQA1 | 0201 | 0201 | 0201 | 0201 |
| TTL | DQB1 | 0502 | 0603 | 050201 | 060301 | CRK | DQB1 | 0201/0202 | 0201/0202 | 0202 | 0202 |
| TTL | DPB1 | 0201 | 1301 | 020102 | 1301 | CRK | DPB1 | 010101 | 110101 | 010101 | 110101 |
| TTL | HLA-A | 1102 | 3303 | 110201 | 330301 | CRK | HLA-A | 2902/4 | 2902/4 | 290201 | 290201 |
| TTL | HLA-B | 51 | 5401 | 510101 | 5401 | CRK | HLA-B | 4403 | 4403 | 440301 | 440301 |
| TTL | HLA-C | 0102 | 0302 | 010201 | 030201 | CRK | HLA-C | 1601 | 1601 | 160101 | 160101 |
| FH6 | DRB1 | 160101 | 1001 | 160101 | 100101 | H0301 | DRB1 | 1302 | 1302 | 130201 | 130201 |
| FH6 | DQA1 | 0101 | 0102 | 010101 | 010201 | H0301 | DQA1 | 0102 | 0102 | 010201 | 010201 |
| FH6 | DQB1 | 0501 | 0502 | 050101 | 050201 | H0301 | DQB1 | 0609 | 0609 | 0609 | 0609 |
| FH6 | DPB1 | 020102 | 0401 | 020102 | 040101 | H0301 | DPB1 | 0501 | 0501 | 0501 | 0501 |
| FH6 | HLA-A | 24 | 2901 | 24020101 | 29010101 | H0301 | HLA-A | 0301 | 0301 | 03010101 | 03010101 |
| FH6 | HLA-B | 0705/6 | 2702 | 070501 | 2702 | H0301 | HLA-B | 1402 | 1402 | 1402 | 1402 |
| FH6 | HLA-C | 0202 | 1505 | 020202 | 150501 | H0301 | HLA-C | 0802 | 0802 | 080201 | 080201 |
| JY | DRB1 | 0404 | 1301 | 0404 | 130101 | OOS | DRB1 | 0101 | 0101 | 010101 | 010101 |
| JY | DQA1 | 0103 | 0301 | 010301 | 030101 | OOS | DQA1 | 0101 | 0101 | 010101 | 010101 |
| JY | DQB1 | 0302 | 0603 | 030201 | 060301 | OOS | DQB1 | 0501 | 0501 | 050101 | 050101 |
| JY | DPB1 | 020102 | 0401 | 020102 | 040101 | OOS | DPB1 | 020102 | 020102 | 020102 | 020102 |
| JY | HLA-A | 020101 | 020101 | 02010101 | 02010101 | OOS | HLA-A | 2601 | 2601/11N | 260101 | 260101 |
| JY | HLA-B | 070201 | 070201 | 070201 | 070201 | OOS | HLA-B | 5601 | 5601 | 5601 | 5601 |
| JY | HLA-C | 0702 | 0702 | 07020101 | 007020101 | OOS | HLA-C | 0102 | 0102 | 010201 | 010201 |
| BM16 | DRB1 | 1201 | 1201 | 120101 | 120101 | SSTO | DRB1 | 0403 | 0403 | 040301 | 040301 |
| BM16 | DQA1 | 0501 | 0501 | 050101 | 050101 | SSTO | DQA1 | 0301 | 0301 | 030101 | 030101 |
| BM16 | DQB1 | 0301 | 0301 | 030101 | 030101 | SSTO | DQB1 | 0305 | 0305 | 030501 | 030501 |
| BM16 | DPB1 | 020102 | 020102 | 020102 | 020102 | SSTO | DPB1 | 0401 | 0401 | 040101 | 040101 |
| BM16 | HLA-A | 0201 | 0201 | 02010101 | 02010101 | SSTO | HLA-A | 3201 | 3201 | 320101 | 320101 |
| BM16 | HLA-B | 1801 | 1801 | 180101 | 180101 | SSTO | HLA-B | 4402 | 4402 | 440201 | 440201 |
| BM16 | HLA-C | 0701 | 0701 | 070101 | 070101 | SSTO | HLA-C | 0501 | 0501 | 050101 | 050101 |
| LH | DRB1 | 0301 | 0404 | 030101 | 0404 | BIN40 | DRB1 | 0404 | 0404 | 0404 | 0404 |
| LH | DQA1 | 0301 | 0501 | 030101 | 050101 | BIN40 | DQA1 | 0301 | 0301 | 030101 | 030101 |
| LH | DQB1 | 0201 | 0402 | 020101 | 0402 | BIN40 | DQB1 | 0302 | 0302 | 030201 | 030201 |
| LH | DPB1 | 010101 | 0501 | 010101 | 0501 | BIN40 | DPB1 | 0301 | 0601 | 030101 | 0601 |
| LH | HLA-A | 2402 | 2402 | 24020101 | 24020101 | BIN40 | HLA-A | 02 | 310102 | 02010101 | 310102 |
| LH | HLA-B | 0802 | 2708 | 080101 | 2701 | BIN40 | HLA-B | 1401 | 4001 | 1401 | 400101 |
| LH | HLA-C | 0102 | 0701/6 | 010201 | 070101 | BIN40 | HLA-C | 03 | 0802 | 030401/0301 | 0802 |
| VOO | DRB1 | 0101 | 030101 | 010101 | 030101 | APD | DRB1 | 1301 | 1301 | 130101 | 130101 |
| VOO | DQA1 | 0101 | 0501 | 010101 | 050101 | APD | DQA1 | 0103 | 0103 | 0103 | 0103 |
| VOO | DQB1 | 0201/2 | 0501 | 020101 | 050101 | APD | DQB1 | 0603 | 0603 | 060301 | 060301 |
| VOO | DPB1 | 020102 | 0401 | 020102 | 040101 | APD | DPB1 | 0402 | 0402 | 0402 | 0402 |
| VOO | HLA-A | 0101 | 0301 | 01010101 | 03010101 | APD | HLA-A | 0101 | 0101 | 01010101 | 01010101 |
| VOO | HLA-B | 0801 | 5601 | 080103 | 5601 | APD | HLA-B | 4001 | 4001 | 400101 | 400101 |
| VOO | HLA-C | 0102 | 0701/06/16 | 010201 | 070101 | APD | HLA-C | 0602 | 0602 | 06020101 | 06020101 |
| AMALA | DRB1 | 1402 | 1402 | 1402 | 1402 | HAR | DRB1 | 0301 | 0301 | 030101 | 030101 |
| AMALA | DQA1 | 0501 | 0501 | 050101 | 050101 | HAR | DQA1 | 0501 | 0501 | 050101 | 050101 |
| AMALA | DQB1 | 0301 | 0301 | 030101 | 030101 | HAR | DQB1 | 0201 | 0201 | 020101 | 020101 |
| AMALA | DPB1 | 0402 | 0402 | 0402 | 0402 | HAR | DPB1 | 040101 | 0401 | 040101 | 040101 |
| AMALA | HLA-A | 021701 | 021701 | 021701 | 21701 | HAR | HLA-A | 0101 | 0101 | 01010101 | 01010101 |
| AMALA | HLA-B | 1501 | 1501 | 15010101 | 15010101 | HAR | HLA-B | 0801 | 0801/5 | 080101 | 080101 |
| AMALA | HLA-C | 0303 | 0303 | 030301 | 030301 | HAR | HLA-C | 0701/6 | 0701/5 | 070101 | 070101 |
Correct calls by Conexio software without editing; Correct calls after minimal manual editing (highlighted); Correct calls after removal of nonspecific sequences (dark highlighted).
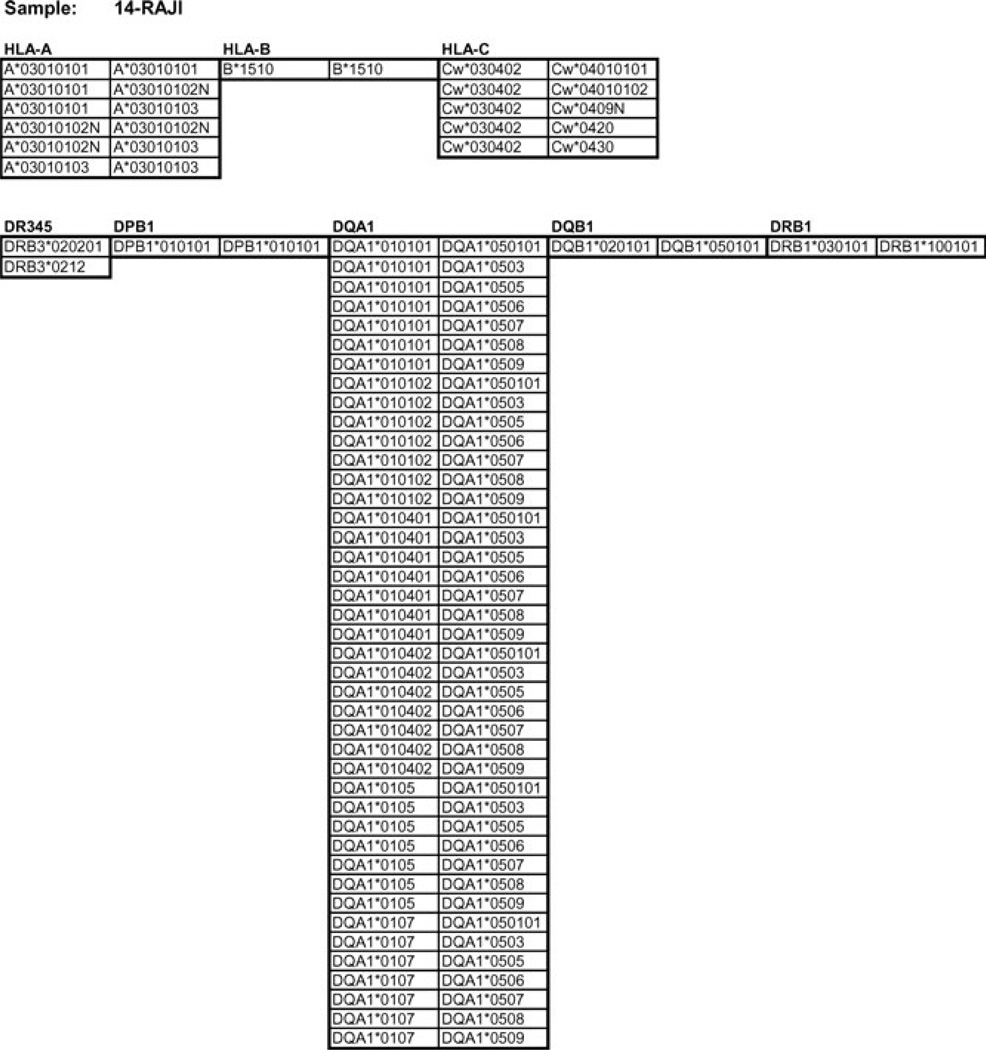
Figure 4.
Conexio genotype assignments for the cell line RAJI report.
The failure to detect A*2601 in this particular 24-sample run was due to a problem with the initial version of the genotyping software in which exon 2 sequence reads that should have been sorted into the primary alignment appeared in the secondary alignment. This problem has been corrected in subsequent versions of the ATF software and we have been able to detect both alleles of this cell line in subsequent runs. Figure S1 (Supporting Information) shows a screen shot of the Conexio genotyping software in which both DBUG HLA-A alleles were correctly assigned. PCR conditions that minimize differential amplification of alleles from genomic DNA should be used, as in all PCR-based typing methods. Also, it is advisable to aim for relatively high numbers of sequence reads per amplicon so that, even if preferential amplification occurs, a sufficient number of sequence reads are recovered to allow assignment of both alleles of a heterozygote by the genotyping software.
A genotype report listing the possible allele assignments (0 mismatches with the database) for all loci tested for each individual sample is generated by the Conexio software. An illustrative example of the genotype assignments for the cell line, RAJI, is shown in Figure 4. Genotype assignments with 0 mismatches of the consensus sequences to the database are shown beneath the locus designation. For those loci with multiple genotype assignments (ambiguity), the polymorphisms that distinguish the primary assignment (first genotype in list) from the other genotypes in the list lie outside of the regions sequenced in this study. The DRB3 locus is on the DRB1*0301 haplotype; the DRB1*1001 haplotype does not carry an additional DRB locus. For DRB1, DQB1, DPB1, and HLA-B, a unique set of two alleles is assigned. For the DRB3 locus, present on the DRB1*03 haplotype, two possible alleles are listed. At the HLA-A locus, the assignment includes several synonymous variants of the A*0301 allele (the first four digits refer to nonsynonymous variants) as well as a very rare ‘null’ variant, A*03010102N, which differs from A*0301 outside the genomic regions sequenced here. Similarly, the three rare alleles listed under HLA-C also differ from the primary assignment, Cw*0401, outside the sequenced region. The long list of possible DQA1 genotypes also reflects differences from the primary assignment, DQA1*0101/DQA1*0501 at polymorphisms that reside outside exon 2. These residual ambiguities can be resolved by additional analyses (i.e. SSP), if desired, or by incorporating additional genomic regions (exons and/or introns) in the 454 sequencing run.
As noted in the Table 2, most genotype assignments were performed by the software without any need for any manual editing, while a subset of samples required some additional intervention (see below). In these samples, the software provides no initial genotype assignment with 0 mismatches. Following manual ‘inactivation’ of rare sequences reads that had not been automatically filtered out, the software provided a genotype assignment with 0 mismatches. In some cases, the manual editing involved ‘trimming back’ bases from the ends of sequence reads that contained mismatches with the consensus sequence. These mismatches reflect pyrosequencing errors that can occur at the ends of reads due to the increased frequency of incomplete extension and carry-forward error that is associated with the incomplete removal of ATP from previous nucleotide incorporation cycles.
Another situation in which manual editing can be required is the genotype assignment of homozygous samples with low numbers of sequence reads; in these cases, the initial version of the software may take relatively rare sequences from the secondary alignment and assign two alleles to this homozygous sample. In all cases, the inspection of the genotyping software output and the manual editing described above, which takes 1–2 min per genotype, yielded the correct genotype assignments. Subsequent versions of the software have addressed these issues and significantly reduce the need for manual editing.
Analysis of mixtures (rare variant detection)
The very high number of sequence reads generated in a typical GS FLX run (300–400K), make possible the detection of rare variant sequences present in the sample. To estimate the sensitivity to detect such sequences, we prepared mixtures of PCR products for exons 2 and 3 of HLA-A and HLA-B from two HLA homozygous cell lines (AMAI, A*680201, B*530101 and SAVC, A*030101, B*70201) in various proportions (1/1, 1/10, 1/100). As can be seen in Table 3, the number of sequence reads for the two alleles of exon 3 of HLA-B were similar in the 1:1 mixture (forward: 1802 vs 1803 and reverse: 1248 vs 1133) as well as the HLA-A exon 2 sequence reads (forward: 1208 vs 1397 and reverse: 816 vs 1051). Since the efficiency of recovering forward and reverse allelic sequence reads for HLA-B exon 3 and HLA-A exon 2 was comparable, these exons were chosen to analyze the 1/10 and 1/100 mixtures. For the HLA-A 1/10 mixture, the ‘rare’ sequence reads (the A*30101 allele) represented 18% of the total forward strand reads and 15% of the reverse strand reads. In the HLA-B 1/10 mixture, the ‘rare’ sequence reads (the B*70201 allele) represented 11% of the total forward strand reads and 10% on the reverse strand reads. The rare variant in 1/100 mixtures could also be readily detected. It was present at a frequency of 0.9% and 1.6% in HLA-A forward and reverse strand reads, respectively, and 2.9% and 2.5% of HLA-B forward and reverse strands, respectively.
Table 3.
Mixtures of homozygous cell-line DNAs (number of sequence reads)
| Dilution ratio 1/1 | Dilution ratio 1/10 | Dilution ratio 1/100 | ||||||
|---|---|---|---|---|---|---|---|---|
| Type | HLA-A | Reads | HLA-A | Reads | Frequency (%) | HLA-A | Reads | Frequency (%) |
| A*680201 | Exon 2 F | 1208 | Exon 2 F | 2201 | Exon 2 F | 1348 | ||
| A*30101 | Exon 2 F | 1397 | Exon 2 F | 471 | 18% | Exon 2 F | 12 | 0.9% |
| A*680201 | Exon 2 R | 816 | Exon 2 R | 1685 | Exon 2 R | 927 | ||
| A*30101 | Exon 2 R | 1051 | Exon 2 R | 304 | 15% | Exon 2 R | 15 | 1.6% |
| Type | HLA-B | HLA-B | HLA-B | |||||
| B*530101 | Exon 3 F | 1802 | Exon 3 F | 4711 | Exon 3 F | 3794 | ||
| B*70201 | Exon 3 F | 1803 | Exon 3 F | 608 | 11% | Exon 3 F | 112 | 2.9% |
| B*530101 | Exon 3 R | 1248 | Exon 3 R | 3295 | Exon 3 R | 2644 | ||
| B*70201 | Exon 3 R | 1133 | Exon 3 R | 363 | 10% | Exon 3 R | 68 | 2.5% |
HLA, human leukocyte antigen.
The blood of certain individuals is chimeric, with residual maternal cells present at very low levels in the child’s circulation or rare fetal cells maintained in the mother’s circulation (19). SCIDS patients often retain circulating maternal cells at very low levels and early detection of maternally derived immunologic cells is important after diagnosis for proper management of the patient. When such patients are recipients of hemapoetic stem cell transplants, characterizing the level of maternal microchimerism is clinically important; exposure to maternal antigens increases the possibility of severe GVHD using unmodified, HLA mismatched related, and unrelated donors in transplantation (20, 21).
Here, we describe the HLA profile of an SCIDS patient, F4R, who was the recipient of an hemapoetic stem cell transplant. The HLA-B and HLA-C types of this patient and his parents, determined by 454 sequencing and the Conexio HLA typing software, based on exon 2 and exon 3 sequence reads, are shown in Table 4. The presence of a ‘third’ HLA-B allele, the nontransmitted maternal allele (B*3512), could be identified in the ‘secondary alignment’ of exon 2 sequence reads. Along with other sequences that represented artifactual variants of the two inherited alleles, there were 11 reverse sequence reads for exon 2 for the B*3512 allele in this secondary alignment, compared with 290 reverse sequence reads for B*3905 and 280 reads for B* 390202 for exon 2 in the primary alignment. A nontransmitted maternal HLA-C* 0401 allele could also be detected in this sample. The HLA-C type of F4R is *0702 homozygous. For exon 3, 1153 copies of a forward sequence read corresponding to the two copies of HLA-C*0702-as well as 10 copies of a forward exon 3 sequence read corresponding to HLA-C*0401 were detected in the primary alignment. In this case, the rare nontransmitted maternal allele is found in the primary rather than the secondary alignment, as in the HLA-B example, because F4R is homozygous at HLA-C so this ‘additional’ allele is the second rather than the third allele. The analysis of the HLA-B and -C sequence reads suggests that the maternal cells in this SCIDS patient’s blood are on the order of 1–2%. A more detailed and systematic analysis by 454 HLA sequencing of microchimerism, examining additional exons and SCIDS patients will be the subject of a subsequent manuscript.
Table 4.
SCIDs patient/recipient and mother and father : HLA-B and HLA-C genotypes
| HLA-B | HLA-C | |||
|---|---|---|---|---|
| F4R | 3905 | 390202 | 07020101 | 07020101 |
| F4M | 3905 | 3512 | 04010101 | 07020101 |
| F4D | 3503 | 390202 | 04010101 | 07020101 |
Alleles B*3512 and C*0401 are the nontransmitted maternal alleles.
HLA, human leukocyte antigen.
Discussion
Allele-level matching for many HLA loci (A, B, C, and DRB1) of donors and recipients is clinically critical for successful HSC transplantation (1–12). In some studies, matching for HLA-DQB1 and DPB1 also has a significant effect on transplant outcome (13–15). Currently, the highest resolution HLA typing is obtained with fluorescent, Sanger-based DNA sequencing using capillary electrophoresis. Even at this level of sequence resolution, ambiguities in the HLA typing data can persist due to multiple polymorphisms shared between alleles and the resultant phase ambiguities when both alleles are amplified and sequenced together. Resolving these ambiguities requires time-consuming approaches such as amplifying and then analyzing the two alleles separately. Clonal sequencing, the analysis of amplicons generated from individual DNA molecules amplified from HLA exons allows the unambiguous sequence determination of the exons and, by comparing these sequence files to an HLA sequence database, the unambiguous determination, in most cases, of the two HLA alleles.
The read lengths achieved by the GS FLX system (avg = 250 bp) allow sufficient overlap for this sequence determination for each exon. The assignment of genotypes at each locus based on the exon sequence data files is performed by a software application developed by Conexio Genomics (Freemantle, AU). A critical aspect of the software is the ability to filter out related sequence reads (pseudogenes and other unwanted HLA genes) that were coamplified along with the target sequence. In most HLA typing methods, such as Sanger sequencing or SSOP typing, these coamplified sequences would generate ‘noise’ and minimize ‘signal’. The software also filters out very rare sequence reads that may have been generated by an error in the initial PCR amplification of the target sequence from genomic DNA, errors in the emulsion PCR, or pyrosequencing errors, as discussed earlier. On the basis of a recent report (22), the pyrosequencing error rate was estimated, in an ultradeep sequencing study of HIV amplicons to be 0.01 in homopolymeric regions (3–5 nucleotides) and 0.002 in nonhomopolymeric regions. The overall error rate was 0.004.
The clonal sequencing property of the 454 GS FLX reveals PCR primer specificity by identifying the sequences of all coamplified genomic regions, in addition to the intended target region. This property is useful in optimizing primer specificity, and allows for the use of generic primers, such as our DRB primers, which amplify DRB3, DRB4, and DRB5, in addition to DRB1, to generate valuable sequence information at multiple loci.
To make the GS FLX system cost-effective for highresolution clinical HLA typing, multiple samples must be analyzed at multiple loci in a single run. The use of MID tags, and multiple picotiter plate regions, makes running 24 or 48 samples analyzed at 7 loci possible and practical (see Table 1). Clearly, larger numbers of samples could be analyzed in a single run using additional MID tags and regions, provided that fewer amplicons per individual were sequenced.
It is the very large number of sequence reads generated in parallel that allows this multiplex analysis of multiple individuals at multiple loci, which also creates the opportunity to detect rare variant sequences. In mixtures of PCR products from two different genomic DNA samples, we were able to reliably detect HLA exon sequences present at a 1/100 dilution (Table 3). The challenge in this application is to filter out related but unwanted sequences, as well as rare sequences containing errors, yet retain and identify the rare allelic variant sequences. HLA sequences are well suited to this kind of analysis because most HLA alleles differ from one another by multiple polymorphisms while the sequences containing errors typically differ from the correct sequence by only one nucleotide.
In addition to the analysis of these cell-line DNA mixtures, the ability to detect rare HLA sequences present in mixtures was demonstrated in the analysis of blood from a SCIDS patient (Table 4). In this patient, the rare nontransmitted maternal allele could be detected along with the inherited maternal and paternal alleles. The potential to analyze chimeric mixtures may have important applications in clinical research (19).
In conclusion, we believe that this high throughput clonal sequencing system can provide cost-effective, reliable, highresolution HLA typing for clinical transplantation, as well as for research studies.
Acknowledgment
We are grateful to Michael Egholm, Birgitte Simen, and Cherie Holcomb for careful review of this manuscript.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1 Sequence of HLA-specific 454 fusion primers (target PCR segment only)
Figure S1 Screenshot of Conexio genotyping software for HLA-A of the cell-line DBUG
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Flomenberg N, Baxter-Lowe LA, Confer D, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104:1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 2.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 3.Kawase T, Morishima Y, Matsuo K, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110:2235–2241. doi: 10.1182/blood-2007-02-072405. [DOI] [PubMed] [Google Scholar]
- 4.Morishima Y, Sasazuki T, Inoko H, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002;99:4200–4206. doi: 10.1182/blood.v99.11.4200. [DOI] [PubMed] [Google Scholar]
- 5.Sasazuki T, Takeo J, Morishima A, Kinukawa N, Kashiwabara H. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from unrelated donor. New Eng J Med. 339:1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 6.Petersdorf EW. HLA matching in allogeneic stem cell transplantation. Curr Opin Hematol. 2004;11:386–391. doi: 10.1097/01.moh.0000143701.88042.d9. [DOI] [PubMed] [Google Scholar]
- 7.Petersdorf EW, Anasetti C, Martin PJ, et al. Limits of HLA mismatching in unrelated hematopoietic cell transplantation. Blood. 2004;104:2976–2980. doi: 10.1182/blood-2004-04-1674. [DOI] [PubMed] [Google Scholar]
- 8.Petersdorf EW, Hansen JA, Martin PJ, et al. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 9.Greinix HT, Fae I, Schneider B, et al. Impact of HLA class I high-resolution mismatches on chronic graft-versus-host disease and survival of patients given hematopoietic stem cell grafts from unrelated donors. Bone Marrow Transplant. 2005;35:57–62. doi: 10.1038/sj.bmt.1704741. [DOI] [PubMed] [Google Scholar]
- 10.Loiseau P, Busson M, Balere ML, et al. HLA Association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13:965–974. doi: 10.1016/j.bbmt.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Maury S, Balere-Appert ML, Chir Z, et al. Unrelated stem cell transplantation for severe acquired aplastic anemia: improved outcome in the era of high-resolution HLA matching between donor and recipient. Haematologica. 2007;92:589–596. doi: 10.3324/haematol.10899. [DOI] [PubMed] [Google Scholar]
- 12.Tiercy JM, Passweg J, van Biezen A, et al. Isolated HLA-C mismatches in unrelated donor transplantation for CML. Bone Marrow Transplant. 2004;34:249–255. doi: 10.1038/sj.bmt.1704569. [DOI] [PubMed] [Google Scholar]
- 13.Horn PA, Elsner HA, Blasczyk R. Tissue typing for hematopoietic cell transplantation: HLA-DQB1 typing should be included. Pediatr Transplant. 2006;10:753–754. doi: 10.1111/j.1399-3046.2006.00558.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaw BE, Gooley TA, Malkki M, et al. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110:4560–4566. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 15.Shaw BE, Marsh SG, Mayor NP, Russell NH, Madrigal JA. HLA-DPB1 matching status has significant implications for recipients of unrelated donor stem cell transplants. Blood. 2006;107:1220–1226. doi: 10.1182/blood-2005-08-3121. [DOI] [PubMed] [Google Scholar]
- 16.Bosch JRT, Grody WW. Review: keeping up with the next generation, massively parallel sequencing in clinical diagnostics. J Mol Diagn. 2008;10:484–492. doi: 10.2353/jmoldx.2008.080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parameswaran P, Jalili R, Tao L, et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams WKM, Nelson JL. Autoimmune disease during pregnancy and the microchimerism legacy of pregnancy. Immunol Invest. 2008;37:631–644. doi: 10.1080/08820130802205886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Small TN, Friedrich W, O’Reilly RJ. Hematopoietic cell transplantation for immunodeficiency disease. In: Blume KG, Forman SJ, Applebaum FR, editors. Thomas’ Hematopoietic Stem Cell Transplantation. 3rd edn. Hoboken: John Wiley and Sons; 2004. pp. 1430–1414. Retention of maternal micro-chimerism may also play a role in chronic inflammatory disease later in life.
- 21.Steves AM. Do maternal cells trigger or perpetuate autoimmune diseases in children? Pediatr Rheumatol Online J. 2007;5:9. doi: 10.1186/1546-0096-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozera G, Abbate I, Bruselles A, et al. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology. 2009;6:15. doi: 10.1186/1742-4690-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]