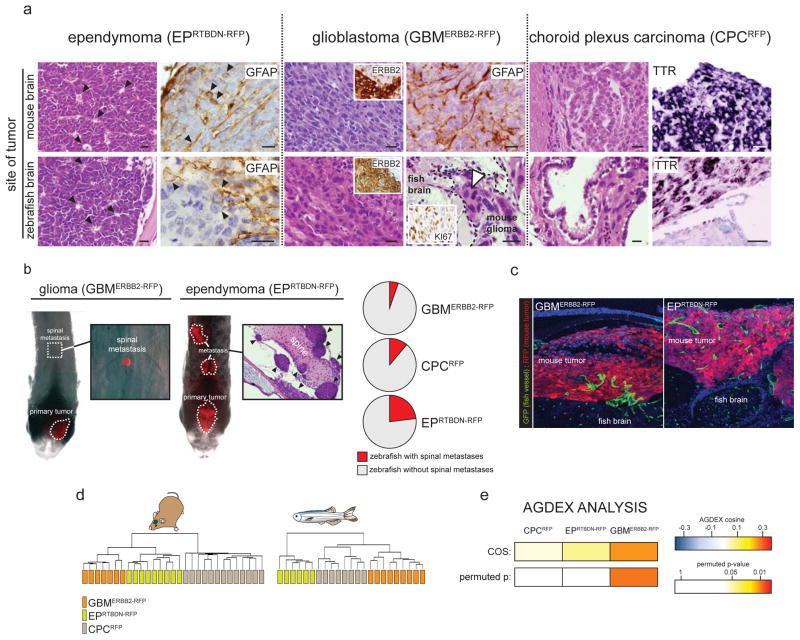
Figure 3. Mouse brain tumors retain key biologic characteristics when xenorafted into zebrafish brains.
(a) Zebrafish harboring tumors were euthanized using 0.04% Tricaine and fixed in 4% paraformaldehyde overnight. Zebrafish were then decalcified using 0.5M EDTA (AMRESCO®) for 5 days, rinsed in phosphate buffered saline, dehydrated and paraffin wax embedded. 5μm sections were stained using hematoxylin and eosin (H&E) or subjected to standard immunohistochemistry using the indicated primary antibodies (GFAP 1:500, Dako rabbit polyclonal Z0334; ERBB2 1:40, Vector mouse monoclonal VP-C380; Ki67, 1:1000, Vector rabbit polyclonal vp-K451). TTR expression was visualized by in situ hybridization using a full length TTR cRNA template (BC032069, generous gift of Dr. Edwin Monuki). Paraffin sections were treated with RNAzip (Ambion, Austin, TX) and de-waxed to water. Probe hybridization was performed at 60°C overnight in standard hybridization buffer. Sections of the same tumors growing in mouse brains were analyzed in parallel for comparison. Arrows in EPRTBDN-RFP H&E and GFAP mark pseudorosettes and GFAP+ tumor cells respectively. Dotted line in bottom right GBMERBB2-RFP H&E demarcates tumor invading normal brain. Arrow indicates mitotic tumor cells (scale bars=15μm). (b) Zebrafish were implanted with the indicated tumors and the entire dorsal brain and spine imaged daily exactly as described in Figure 1 and 2. Arrows in H&E of EPRTBDN-RFP tumors mark individual metastatic deposits. Pie charts report the proportion of zebrafish in which the indicated tumors metastasized. (c) Fli1:eGFP zebrafish harboring tumors were euthanized fixed and decalcified as described above. Brains containing tumor were then cryo-protected in 30% sucrose for 2 days and frozen in tissue freezing media (TBS®). 12μm sections were counter stained using DAPI containing hard set mounting media and imaged by confocal microscopy with 488nm and 594nm filters. (d) Total RNA was extracted from the indicated brain tumors grown in mouse and zebrafish brains. Microarray gene expression profiles were generated using Affymetrix 430v2 arrays and subject to unsupervised hierarchical clustering exactly as described7. Transcriptomes of each tumor type grown in mouse and human brains were directly compared using the Agreement of Differential Expression (AGDEX) algorithm exactly as described was used7.