Figure 1.
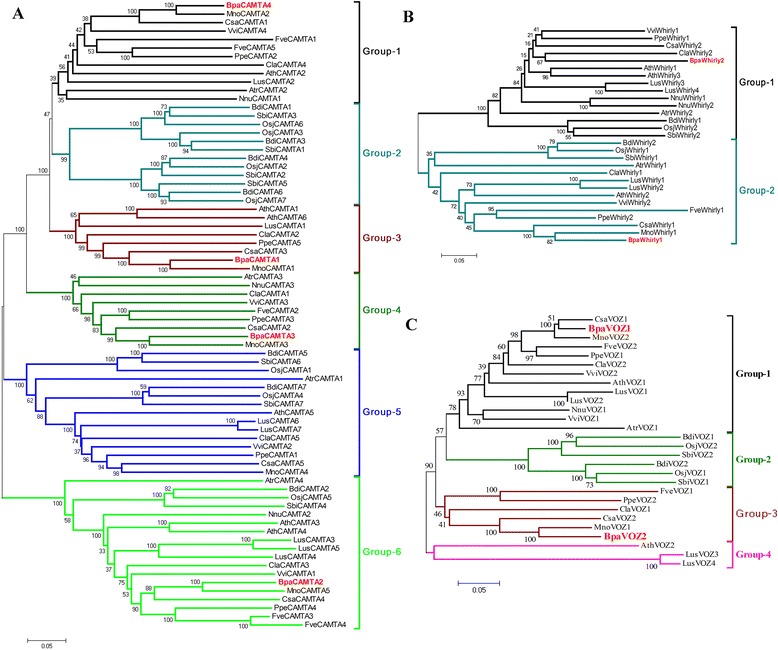
Evolutionary relationships revealed by phylogenetic analysis of CAMTA, Whirly and VOZ family. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1500 replicates) was shown next to the branches. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and were in the units of the number of amino acid differences per site. The analysis involved 74 CAMTAs (A), 29 Whirlys (B) and 27 VOZs (C) amino acid sequences and their correspondent accession numbers were list in Additional file 4: Table S6. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA5.
