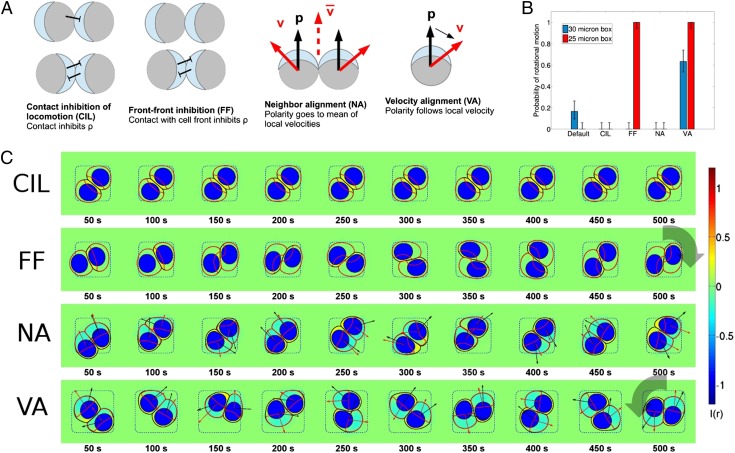
Fig. 3.
Polarity mechanisms can control persistent rotational motion. (A) We implement four distinct mechanisms for cell polarity. In CIL cell contact locally generates I(r), inhibiting ρ. In FF, I(r) is generated when a cell contacts another cell’s front. Here, ρ is indicated by light blue at the front of the cell. In the NA and VA mechanisms, we introduce an auxiliary polarity vector with its own equation of motion. NA orients to the local average of velocities in its neighborhood with a timescale Torient; this resembles flocking-inspired models. VA orients along the cell’s velocity with a timescale Torient. In both of these models the cell’s chemical polarity is then biased to orient the front of the cell (high ρ) with the polarity . There is also a noise term; fluctuates around its target direction. Details of the implementation of these models are described in SI Appendix. (B) PRM probability is controlled by polarity mechanisms; error bars are 68% confidence intervals by the method of ref. 50. “Default” indicates the minimal model with no polarity mechanism. (C) Representative time traces are shown for different polarity mechanisms. Micropattern size Lmicro = 25 μm. As above, cell boundary is shown as a black line, ρ(r) by its contour (in red), the nucleus by a blue shape, and I(r) by the color map. In VA and NA mechanisms cell velocity is shown as a red arrow, and the polarity vector as a black arrow. See also Movies S3–S11 and SI Appendix, Fig. S3.