Significance
Exosomes have been a subject of great interest in recent years, especially in the context of the microRNAs (miRNAs) that they contain. Exosome-mediated miRNA transfer between cells has been proposed to be a mechanism for intercellular signaling and exosome-associated miRNAs in biofluids have been suggested as potential minimally invasive biomarkers for multiple human disease states. Remarkably, we show here that most exosomes derived from standard preparations do not harbor many copies of miRNA molecules. These findings suggest a reevaluation of current models of the mechanism of exosome-mediated miRNA communication and indicate that stoichiometric analysis will be valuable for the study of other populations of extracellular vesicles and their associated RNAs as well.
Keywords: microvesicle, circulating
Abstract
Exosomes have been proposed as vehicles for microRNA (miRNA) -based intercellular communication and a source of miRNA biomarkers in bodily fluids. Although exosome preparations contain miRNAs, a quantitative analysis of their abundance and stoichiometry is lacking. In the course of studying cancer-associated extracellular miRNAs in patient blood samples, we found that exosome fractions contained a small minority of the miRNA content of plasma. This low yield prompted us to perform a more quantitative assessment of the relationship between miRNAs and exosomes using a stoichiometric approach. We quantified both the number of exosomes and the number of miRNA molecules in replicate samples that were isolated from five diverse sources (i.e., plasma, seminal fluid, dendritic cells, mast cells, and ovarian cancer cells). Regardless of the source, on average, there was far less than one molecule of a given miRNA per exosome, even for the most abundant miRNAs in exosome preparations (mean ± SD across six exosome sources: 0.00825 ± 0.02 miRNA molecules/exosome). Thus, if miRNAs were distributed homogenously across the exosome population, on average, over 100 exosomes would need to be examined to observe one copy of a given abundant miRNA. This stoichiometry of miRNAs and exosomes suggests that most individual exosomes in standard preparations do not carry biologically significant numbers of miRNAs and are, therefore, individually unlikely to be functional as vehicles for miRNA-based communication. We propose revised models to reconcile the exosome-mediated, miRNA-based intercellular communication hypothesis with the observed stoichiometry of miRNAs associated with exosomes.
Exosomes have been classically described as 40- to 100-nm vesicles (1, 2) that are secreted by a broad range of cell types and have been identified in diverse body fluids (e.g., plasma, saliva, lymph, ascites, semen, amniotic fluid, cerebrospinal fluid, etc.) (ref. 3 and references therein). More recently, microRNAs (miRNAs) have been reported to be present in exosome preparations (4), suggesting that these vesicles may function as a vehicle for intercellular miRNA transfer [“a message in a bottle” (5)] and a mode of intercellular communication (6). This hypothesis has been tantalizing, given that miRNAs bind to and repress the activity of specific target mRNAs, and it is estimated that >60% of all mRNAs are regulated by miRNAs (7). In addition, exosomes typically display cell surface proteins derived from their cell of origin, which can be recognized by cell surface receptors (e.g., proteoglycans) and internalized by recipient cells (8), resulting in transfer of the exosome contents. Intercellular transfer of miRNAs through exosomes would bypass recipient cell transcriptional controls (9), providing a relatively direct means of regulation. Studies delivering purified exosomes to recipient cells have reported transfer of miRNAs in experimental settings (10–12), and the use of exosomes as small RNA delivery vehicles is being studied as a potential therapeutic strategy (13, 14).
Although the possibility of exosome-mediated miRNA transfer as a mode of intercellular communication is an attractive concept (6, 15, 16), current mechanistic models lack detail, and the physiologic significance of this paradigm is not yet established (17). Quantitative evaluation of key components is fundamental to testing the validity of any model (18), and knowledge of the stoichiometry of miRNAs and exosomes (i.e., how many molecules of a given miRNA are carried by an exosome) would provide insight into the requirements and limits of miRNA-based intercellular communication. This question has historically been challenging to address, because exosomes are subdiffraction limit particles and therefore, cannot be directly enumerated by light microscopy or flow cytometric methods (19). Although exosomes are typically visualized by electron microscopy (EM) (20), this approach is nonquantitative because of the variation introduced by the complex sample preparation process. One method that can circumvent these limitations is nanoparticle tracking analysis (NTA). NTA quantifies particles under 300 nm by visualizing the light scattering generated by the particles and using it to track them during Brownian motion (19).
In this study, we set out to determine the degree to which extracellular biomarker miRNAs are associated with exosomes prepared from the plasma of prostate cancer patients given that exosomes have been proposed as a significant packaging mechanism for miRNAs released from cancer cells (21–25). We found that only a small minority (median = 2.5%) of the extracellular miRNA content of plasma is associated with the exosomal fraction prepared by standard differential centrifugation methods, prompting us to use quantitative methods to more directly determine the stoichiometry of miRNAs and exosomes from cancer patient plasma as well as diverse other biological sources. We show here that the stoichiometry of a given miRNA and exosome is consistently much less than one, even for the most abundant miRNAs identified in our exosome preparations. Thus, most individual exosomes in standard preparations do not carry biologically significant numbers of miRNAs. These results have implications for exosome-mediated miRNA communication (6, 15) and indicate that current mechanistic models for this phenomenon may need to be revised to be reconciled with the observed miRNA–exosome stoichiometry.
Results
Standard Exosome Preparations Recover Only a Minority of miRNAs from Cancer Patient Plasma.
Multiple independent studies have reported that exosomes contain miRNA, and exosomes have been proposed to be treasure chests for biomarker applications (22, 26). However, we and other groups have found that, in healthy individuals, many cell-free circulating miRNAs are present in soluble form and complexed with proteins, such as Argonaute2 (Ago2) (27, 28). Although circulating miRNA biomarkers for cancer have been observed in exosome preparations (22, 29), a quantitative analysis of the physical state (i.e., exosome-associated vs. soluble) of cancer-associated miRNAs has not been reported. Given that cancer cells are reported to abundantly shed exosomes, we hypothesized that cancer-associated circulating miRNAs (and consequently, those most suitable as biomarkers) would be packaged primarily in exosomes.
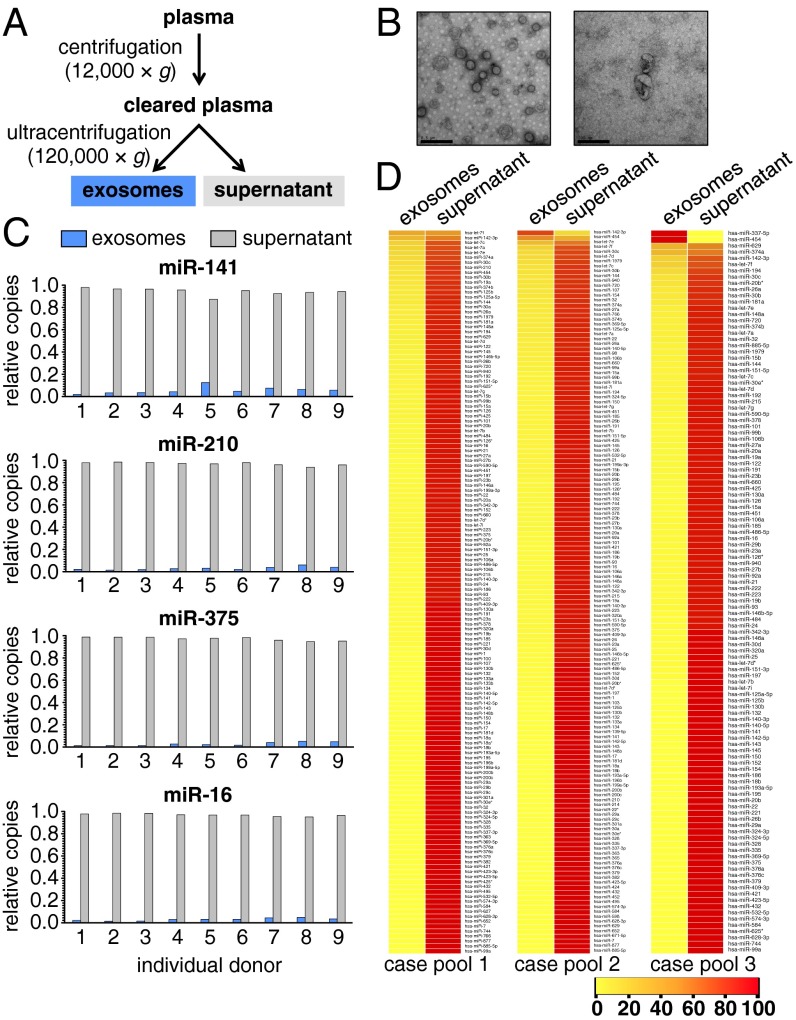
To determine the physical state of cancer-associated circulating miRNAs, we chose to examine the abundance of three established circulating prostate cancer biomarkers [hsa-miR-141 (30), -210 (31), and -375 (22, 31)] and a broadly expressed control miRNA found to be nondiagnostic in plasma [hsa-miR-16 (30)] in exosome preparations from plasma collected from patients with metastatic prostate cancer (n = 9). Plasma samples were fractionated by differential centrifugation, a typical method used in the field to prepare exosomes (Fig. 1A and SI Materials and Methods) (20, 32, 33). Transmission EM (TEM) was used to confirm the recovery of vesicles that were consistent with exosomes by size and morphology (Fig. 1B). miRNA was extracted from aliquots of both the supernatant and exosome fractions, and the abundance of the four miRNAs listed above was quantified by real-time PCR. The exosome fraction contained little miRNA relative to the supernatant (mean values ranging from 2.7% for miR-375 to 5.6% for miR-141) (Fig. 1C and Table S1). Comparable results were obtained from the analysis of exosome fractions prepared from patient serum (n = 3) (Fig. S1) and fresh (i.e., never frozen) plasma (n = 6) (Fig. S2), indicating that our results were not specific to plasma and not an artifact of freezing. To determine if the observed distribution of cancer biomarker miRNAs was representative of the global profile of miRNAs in plasma from these patients, we measured the relative abundance of 375 individual miRNAs in the plasma fractions using real-time PCR array analysis of pooled samples (n = 3 individuals per pool in three pools) (Fig. 1D). On average, we detected 137 miRNAs in the pools by real-time PCR, and only 2.5% (median) (Table S2) of the miRNA content of these miRNAs was present in the exosome fractions.
Fig. 1.
Exosome preparations from cancer patient plasma have low miRNA abundance. (A) Differential centrifugation workflow used to prepare exosome fractions. (B) Representative TEM image of exosome preparations. (Scale bars: Left, 0.5 μm; Right, 200 nm.) (C) Relative abundance of selected biomarker (miR-141, -210, and -375) and nonbiomarker control (miR-16) miRNAs in plasma exosome fractions (blue bars) and postultracentrifugation supernatant (gray bars) across prostate cancer patient plasmas (n = 9). (D) Heat map of relative quantity values (relative percentage of abundance) for miRNAs detected by real-time PCR array profiling of exosome and supernatant fractions derived from prostate cancer patient plasmas (n = 3 pools composed of N = 3 individuals each). miRNAs are ranked (top to bottom) according to relative proportion of each miRNA in exosome vs. supernatant fractions. Consistent with our previous study using healthy donor plasma, a small but notable minority of miRNAs was enriched in the exosome fraction, including the hematopoietic-specific miRNA miR-142–3p.
Exosomes from Diverse Biologic Sources Contain Less than One Copy, on Average, of Their Most Abundant miRNAs.
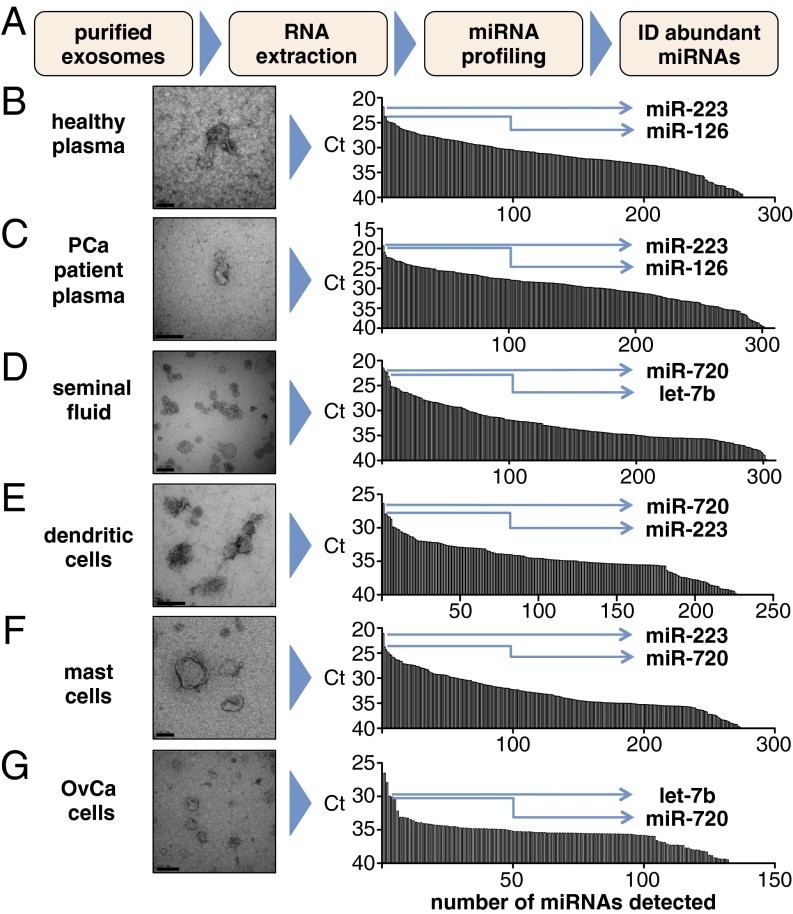
Although we observed that the proportion of total circulating miRNA associated with exosome fractions was small, we hypothesized that individual exosomes may still carry a significant number of miRNA molecules and thereby, provide potency for intercellular communication (34). To test this hypothesis, we sought to quantify the number of miRNA molecules (of a given sequence) that are contained within an exosome. To provide data that would be broadly representative, we prepared exosomes from diverse sources, including different human biofluids (healthy donor plasma, n = 3 donors; prostate cancer patient plasma, n = 3 donors; healthy donor seminal fluid, n = 3 donors), as well as in vitro sources (human dendritic cells, n = 3 donors; ovarian cancer cells, n = 3 preparations; mast cells, n = 2 preparations) to also evaluate more homogenous exosomes (i.e., produced by single-cell types) (4). Exosomes were prepared using typical ultracentrifugation-based protocols (SI Materials and Methods). TEM was used to confirm that the protocols yielded vesicles that were consistent with exosomes by size and morphology (Fig. 2).
Fig. 2.
Identification of abundant miRNAs in exosome preparations from diverse sources. (A) Schematic of the workflow used to identify (ID) abundant miRNAs. (B–G) TEM images of exosomes prepared from (B) healthy (donor) plasma, (C) prostate cancer (PCa) patient plasma, (D) healthy donor seminal fluid, (E) dendritic cells (in vitro differentiated Langerhans cells), (F) mast cells (HMC-1 cell line), and (G) ovarian carcinoma (OvCa) cells (2008 cell line). Real-time PCR array profiling results for all detected miRNAs [sorted by abundance as represented by cycle threshold (Ct) on a reverse y axis] are displayed in Right for the corresponding samples in Left. Each bar represents a different miRNA. miRNAs selected for absolute copy number quantification from the top five most abundant (as defined by the lowest Ct values) are indicated by arrows. Primary miRNA profiling data are also presented in Table S4. (Scale bars: B, D, and F, 100 nm; C, E, and G, 200 nm.)
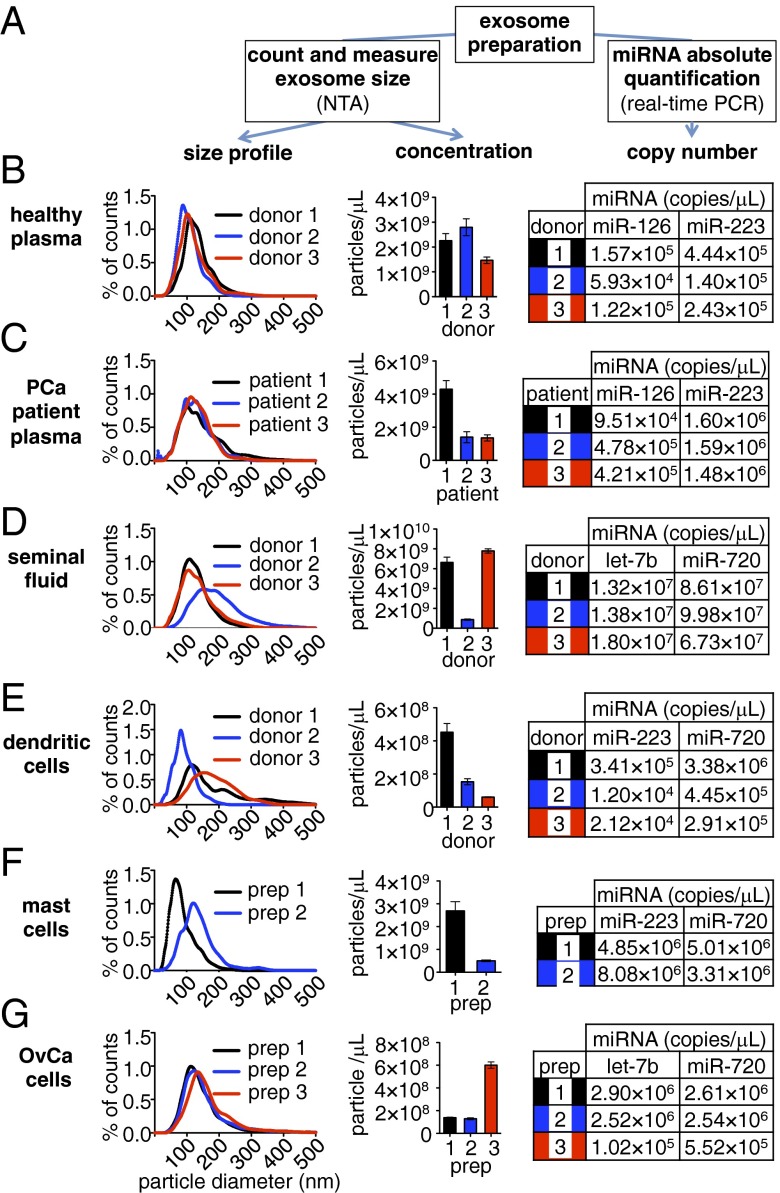
We then measured the number of exosomes in each of the samples using NTA (Fig. 3 and SI Materials and Methods). Exosome concentrations ranged over two orders of magnitude from 6.06 × 107 exosomes/μL in one dendritic cell-derived preparation to 7.79 × 109 exosomes/μL in a semen exosome preparation. Size distribution profile analysis of the NTA data confirmed populations with particle sizes consistent with exosomes, with an average modal size ranging from 92 (for mast cell exosomes) to 122 nm (for seminal fluid exosomes) (Fig. 3), reflective of their appearance by EM (Fig. 2).
Fig. 3.
Quantification of exosome number, size distribution, and miRNA content. Aliquots of exosome preparations described in Fig. 2 were counted and sized by NTA. In parallel, total RNA was extracted from additional exosome aliquots, and the concentrations of abundant exosomal miRNAs identified in Fig. 2 were determined by real-time PCR. (A) Workflow to determine the size distribution, exosome concentration, and absolute quantification of miRNAs in each exosome sample. (B–G) Exosome size distribution histograms (representing the percentage of total counts found within each 1-nm-sized bin), total particle concentrations, and miRNA concentrations in exosome preparations for (B) healthy (donor) plasma (n = 3 donors), (C) prostate cancer (PCa) patient plasma (n = 3 patients), (D) healthy donor seminal fluid (n = 3 donors), (E) dendritic cells (in vitro differentiated Langerhans cells; n = 3 donors), (F) mast cells (HMC-1 cell line; n = 2 preparative replicates: preparation 1 from serum-free conditioned medium and preparation 2 from exosome-depleted serum containing medium), and (G) ovarian carcinoma (OvCa) cells (2008 cell line; n = 3 preparative replicates). Values represent the concentrations in each exosome preparation (i.e., not the concentration in the crude starting specimen).
Although the accuracy of exosome counts derived from NTA has been independently established (19, 35), we empirically validated this approach by comparing NTA-derived counts with those produced using an independent method of quantification: comparative fluorescence microscopy analysis (SI Materials and Methods and Table S3). This method is based on the quantification of dye-labeled exosomes relative to fluorescent beads of a comparable size. Results produced by NTA were similar to those obtained by comparative fluorescence in triplicate analysis of the same exosome sample (8.9 × 1010 vs. 1.3 × 1011 exosomes/mL, respectively; P = 0.5619).
To identify the most abundant miRNAs in each exosome type, we extracted total RNA from aliquots of each sample, pooled the samples corresponding to a given exosome type, and analyzed each pool by real-time PCR array analysis. Abundant miRNAs were identified by ranking cycle threshold data in ascending order (Fig. 2 and Table S4). We then performed absolute quantification of two of five miRNAs displaying the lowest cycle thresholds for each of the individual (nonpooled) samples using a standard real-time PCR-based absolute quantification protocol for miRNAs (Fig. 3 B, Right, C, Right, D, Right, E, Right, F, Right, and G, Right) (36).
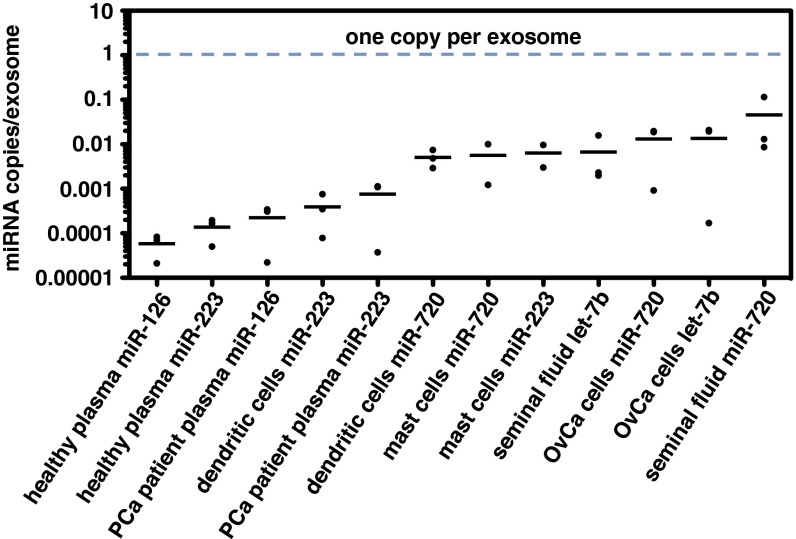
Using the data derived from the above experiments, we determined the ratio of miRNA molecules to exosomes for each sample. In all of the samples examined, the ratio of miRNA molecules for a given miRNA to the number of individual exosomes was substantially lower than one (Fig. 4 and Table S5). Across all samples, we observed an average of one copy of miRNA detected per 121 exosomes, ranging from one copy per 9 exosomes (miR-720 in seminal fluid exosomes) to one copy per 47,162 exosomes (miR-126 in healthy donor plasma exosomes). We found this result surprising, because it implied that most exosomes would not contain any copies of abundant miRNAs.
Fig. 4.
Abundant miRNAs in exosomes are present at much less than one copy per exosome. Dot plot of miRNA copies per exosome determined for each exosome source (based on data collected like in Fig. 3) and sorted by mean value (bar). Numeric values are presented in Table S5. OvCa, ovarian carcinoma; PCa, prostate cancer.
We sought to exclude the possibility that confounding technical factors may have impacted our results, resulting in an underestimate of miRNA abundance and/or an overestimate of exosome number. First, we sought to determine whether miRNA amplification efficiency might be reduced in the exosome samples (e.g., because of the copurification and enrichment of a reverse transcription or PCR inhibitor in the specimens). This reduced amplification efficiency could artificially lower our estimation of miRNA abundance. However, there was no statistically significant difference (P = 0.1038) in the measured values of the spiked-in synthetic cel-miR-39 miRNA oligoribonucleotide control (SI Materials and Methods) (36) in RNA extracts prepared from exosomes vs. those prepared from the postultracentrifugation supernatant. This result indicated that there was no overall reduction in sensitivity for miRNA detection in exosome samples relative to the supernatant (Materials and Methods and Fig. S3). Likewise, there was no difference (P = 0.7587) between exosome and supernatant samples with respect to measured values of synthetic oligoribonucleotide controls (UniSp6) (SI Materials and Methods) that were spiked into the reverse transcription reaction mixtures (Fig. S3), consistent with no difference in the abundance of potential reverse transcription or PCR inhibitors.
Second, we considered the possibility that our exosome preparations may have been contaminated by cosedimentation of other particles endogenously present in the crude samples. These particles could be miscounted by NTA as exosomes, resulting in an overestimate of exosome abundance and therefore, reducing our estimate of miRNA copies per exosome. However, the diverse nature of specimen types (ranging from cell culture supernatants to body fluids) and the use of different purification methods (SI Materials and Methods) make a consistent and abundant contaminant unlikely. Furthermore, TEM images are not consistent with such a predominant contaminant. Even in the case of exosomes prepared from a complex fluid, like plasma (Fig. 2 B and C), although there were additional background particles observed on TEM, they were predominantly very small (∼10 nm; possibly representing HDL) and generally below the size limit of detection of NTA. In contrast, exosomes purified from mast and ovarian cancer cells in culture (Fig. 2 F and G) did not display such additional small particles but still yielded far less than one miRNA molecule per exosome. In addition, the modal distribution of the sizes of particles observed by NTA is consistent with expectations of exosomes and TEM observations.
Third, we addressed the possibility that our results could be the product of inaccurate miRNA quantification caused by the dependence of real-time PCR on standard curves (37). We recently described (38) a droplet digital PCR (ddPCR) -based approach for absolute quantification of miRNAs from biofluid samples. This end point method is independent of standard curves, shows higher precision for the quantification of low-abundance miRNA targets, and is resistant to residual PCR inhibitors (39). To determine whether ddPCR would confirm our overall findings, we used ddPCR and real-time PCR side by side to quantify miR-223 from healthy donor plasma exosomes as well as let-7b and miR-720 from ovarian cancer cell exosomes (Fig. S4). Quantification of miR-223 and let-7b differed by less than 1% and 27% between the two methods, respectively, which were not statistically significant (P = 0.8861 and P = 0.1696, respectively). miR-720 measured in ovarian cancer cell exosomes was found to be present at 70% lower abundance when measured by ddPCR vs. real-time PCR (2.71 × 10−4 vs. 9.16 × 10−4 copies per exosome, respectively; P = 0.0040), indicating that this miRNA may be even more rare in exosomes than estimated by real-time PCR.
Discussion
The discovery that exosomes are associated with miRNAs has spawned great interest in these vesicles as potential vehicles for miRNA-based intercellular communication as well as a source of diagnostic extracellular miRNA biomarkers. However, quantitative analyses to inform our mechanistic understanding of exosome-mediated miRNA communication and guide our approach to the development of exosome-based molecular diagnostics have been lacking. Exosomes are several orders of magnitude smaller than cells (i.e., if we model both as spheres with exosome diameter = 100 nm and cellular diameter = 10 μm, an exosome would be estimated to contain 0.0001% of the cellular volume). Although this very small size implies that exosomes have a very limited capacity to sample the diverse RNA repertoire, some studies have reported that certain miRNA sequences are enriched in exosomes relative to their cells of origin (4, 10). Such enrichment could yield exosomes containing selected miRNAs at much higher copy numbers than would be expected from random sampling. However, our results show that even abundant miRNAs are present at far less than one copy per exosome, indicating that most exosomes from standard preparations are devoid of the most common sequences. This finding seems to be generalizable, because the same conclusion was reached across six different sources of exosomes derived from various body fluids as well as conditioned media from in vitro cell cultures.
We considered it important to exclude the possibility of technical inaccuracy of our miRNA and/or exosome quantification methods, although such error would have to be on the scale of orders of magnitude to change the overall conclusions. Therefore, we validated both our miRNA and exosome quantification methods using orthogonal approaches (i.e., ddPCR and membrane labeling/fluorescence microscopy, respectively) and confirmed empirically that a difference in neither PCR amplification efficiency nor exosome quantification could explain our results. We also ruled out the possibility that freezing and subsequent thawing of exosome samples could be a confounding variable, which is consistent with an independent report confirming the stability of exosomes and exosomal RNA under conditions of freezing–thawing (40).
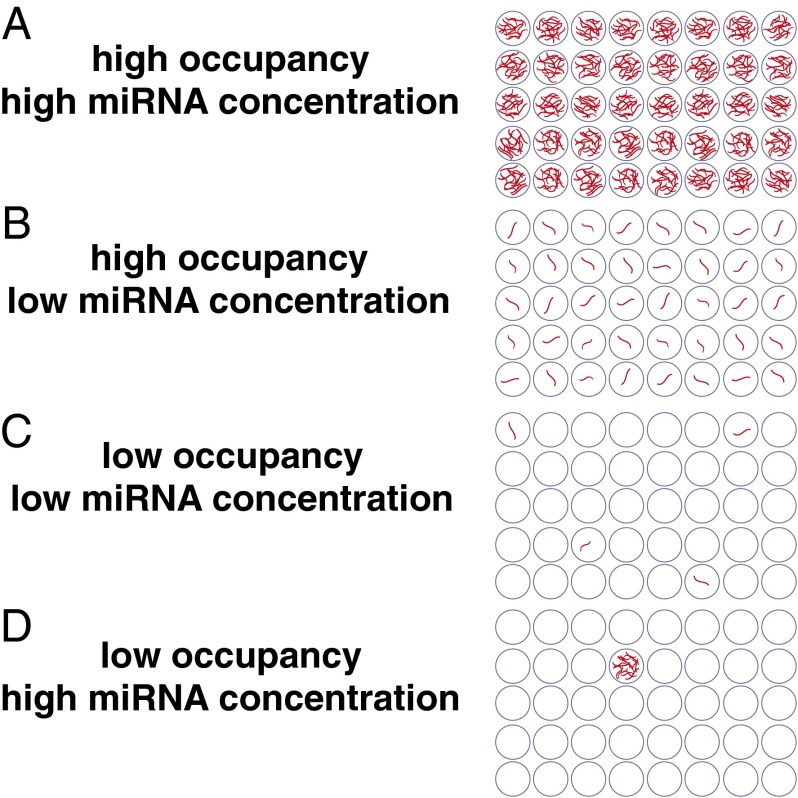
Multiple reports have implicated exosomes as a vehicle for cell–cell communication through carriage and transfer of miRNAs between cells. In general, studies of this particular function have typically been done with a large excess of exosomes, and whether it is feasible for endogenous exosomes to be functional miRNA transfer vehicles in native physiological settings is not yet well-established. Our observed stoichiometry would suggest that most individual exosomes are unlikely to be functional for miRNA communication. Our results refute both a high-occupancy/high-miRNA concentration model (Fig. 5A) and a high-occupancy/low-miRNA concentration model (Fig. 5B). However, we propose that our data are consistent with two alternative models: (i) a low-occupancy/low-miRNA concentration model (Fig. 5C), in which a small fraction of exosomes carries a low concentration of miRNA, or (ii) a low-occupancy/high-miRNA concentration model (Fig. 5D), in which there are rare exosomes in the population carrying many copies of a given miRNA. Both of these models would yield an average stoichiometry of less than one miRNA per exosome. If exosome uptake is a selective and infrequent event for a given cell, the low-occupancy/high-concentration occupancy model would seem to be the most likely to support physiologic exosome-mediated communication through conventional RNA-induced silencing complex-mediated targeting of mRNAs. Our results, therefore, suggest that additional study of potentially miRNA-rich subpopulations of exosomes may be important in investigating intercellular miRNA-based communication. However, if cellular uptake of exosomes is rapid, low-concentration/low-occupancy miRNA may accumulate within the cell in functionally sufficient quantities. Such rapid uptake has been observed in macrophages, which can internalize an equivalent of their cell surface in pinocytic vesicles every 33 min (41). In addition, it is worth noting that both low-occupancy models could be compatible with recently proposed nonconventional activities of miRNAs that presumably require much lower concentrations of miRNA delivery than conventional RNA-induced silencing complex-mediated mRNA targeting. Such activities include the elicitation of cellular responses through binding of Toll-like receptors (42, 43) as well as potential effects of small RNAs on DNA transcription and/or epigenetic states (44).
Fig. 5.
Stoichiometric models for exosome miRNA content. (A) High-occupancy/high-miRNA concentration model, in which the number of molecules of an individual miRNA sequence would far exceed the number of exosomes. (B) High-occupancy/low-miRNA concentration model, in which the concentration of miRNA is lower, but most exosomes contain the miRNA. If the number of exosomes exceeds the number of copies of a given miRNA (as observed in our study), the miRNA molecules may be (C) distributed throughout the population in a low-occupancy/low-miRNA concentration distribution or (D) amassed in rare exosomes in a low-occupancy/high-miRNA concentration distribution.
Exosomes isolated from plasma and serum have also been reported as potential sources of miRNA biomarkers for minimally invasive disease diagnostics. We observed that only a small minority (<3%) of cancer-associated biomarker miRNAs were recovered with classical exosomes isolated using gold standard differential ultracentrifugation-based methods. Although this result does not directly address the diagnostic use of these exosomes, it indicates that the majority of these established biomarkers is present in plasma and serum in other physical forms. Future rigorous studies will be required to determine the diagnostic use of classical exosomes relative to extracellular vesicle preparations recovered by alternate methods as well as other physical and biochemical fractions of patient plasma. Larger vesicles, such as microvesicles and oncosomes, have also been reported to contain miRNAs as well as larger RNAs (45, 46), and our protocols were not designed to recover such vesicles. It is possible that such larger classes of extracellular vesicles may carry physiologically significant numbers of miRNA molecules and function as an alternative vehicle for miRNA-based intercellular communication as well as a source of biomarkers.
In addition, whereas our study focused on miRNAs, it is possible that other classes of RNA commonly measured in such biomarker studies, such as mRNAs (4, 45), may be packaged differently than miRNAs. Future studies will be required to understand the relationship of other classes of RNA with different classes of extracellular vesicles.
In conclusion, our study of the stoichiometry of extracellular vesicles and the miRNA cargo that they carry provides a fundamental approach and a quantitative mechanistic framework to delineate the functional boundaries of extracellular vesicle-mediated communication. Our data warrant the specific quantitative evaluation of exosomes that have been observed to be biologically active in other settings [e.g., glioblastoma (45), placental immunity to viral infections (47), etc.] and thought to exercise their effects through miRNAs, because the relevant exosome–miRNA stoichiometries may contrast with those reported here and provide valuable insight into their mechanism and physiologic relevance.
Materials and Methods
Human Specimens.
Written informed consent was obtained from each donor, and the study was performed with Institutional Review Board (IRB) approval as specified below.
Plasma.
The research was approved by the University of Washington and Fred Hutchinson Cancer Research Center IRBs. Venous blood samples from individuals with metastatic prostate cancer and prostate cancer-negative controls were collected and centrifuged for 15 min at 1,300 × g at 4 °C. Plasma was aspirated and stored at −80 °C.
Cord blood collection and dendritic (Langerhans) cell derivation.
Approval for the study was given by the Seattle Biomedical Research Institute IRB. Isolated CD34+ cells were differentiated as independently published (48, 49). Purified clusters were cultured at 5 × 105 cells/mL for 3 d, after which culture medium was used for exosome isolation (see below).
Semen collection and preparation.
Approval for the study was given by the University of Washington IRB. Ejaculates from healthy HIV-negative men were mixed with 3 mL RPMI medium and kept on ice until processing.
Isolation of Exosomes.
From plasma.
Plasma or serum (1 mL) was added to 1 mL ice-cold PBS and centrifuged at 12,000 × g for 45 min at 4 °C. The cleared dilute plasma was then aspirated, added to 3 mL ice-cold PBS, and centrifuged at 120,000 × g for 70 min. Supernatants were gently decanted, and exosomes were resuspended in PBS.
From Langerhans cell-conditioned medium and semen.
Cells were cultured for 3 d in exosome-depleted media. Cells were removed by centrifugation at 1,000 × g for 10 min, and debris was removed by centrifugation at 2,400 × g for 30 min followed by syringe filtration. Exosomes were then purified by ultracentrifugation over a sucrose cushion using a method adapted from ref. 50.
From human ovarian carcinoma cells.
The 2008 ovarian cancer cells (51) were grown in DMEM with 10% (vol/vol) FBS. Cultures were then washed in triplicate and grown in serum-free medium for 48 h. These media were collected, centrifuged at 500 × g for 10 min, 0.22-µm syringe filtered, and then, ultracentrifuged as above for preparation of plasma exosomes.
From mast cells.
Exosomes were prepared from the HMC-1 cell line essentially as described in ref. 4.
EM.
Exosomes were processed for visualization by TEM as previously described (20).
RNA Isolation.
Total RNA was isolated from all samples using the miRNeasy Kit (Qiagen) as described in ref. 36.
Individual Real-Time PCR Assays.
Individual miRNAs were detected by real-time PCR as previously described (30, 36).
ddPCR.
Samples were analyzed using our recently described method (38).
miRNA Profiling.
Samples were profiled using miRNA Ready-to-Use PCR, Human Panel I, V2.M (Exiqon). The limit of quantification and the PCR efficiency for each miRNA assay were determined as described in ref. 27.
NTA.
Samples were loaded into the assembled sample chamber of a NanoSight LM10; 60-s video images were acquired by a Hamamatsu C11440 ORCA-Flash 2.8 digital camera and analyzed by NanoSight NTA 2.3 software.
Statistical Analysis.
Tests and parameters are indicated in the figures. All statistical analyses were performed using Prism 5.0c software.
Details of sample collection, processing, experimental procedures, and data analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Fred Hutchinson Cancer Research Center Electron Microscopy Shared Resource (B. Schneider, S. MacFarlane, and S. Knecht) and the University of Washington Keck Microscopy Facility for assistance in preparing and visualizing samples; R. Parkin, J. Noteboom, and D. Gonzales for assistance with specimen collection; J. Guenthoer, S. Freggario, J. Yan, D. Kuppers, and W. Spangenberg for advice and helpful discussion; D. Adair for administrative support; and G. Powell for research coordination. We especially thank the volunteers who provided specimens for this study. Fig. 5 art was prepared by Mesa Schumacher, Information Art and Design. This work was supported by Canary Foundation/American Cancer Society (ACS) Postdoctoral Fellowship for the Early Detection of Cancer PFTED-09-249-01-SEID (to J.R.C.), the Hillcrest Committee of Southern Oregon (J.R.C. and J.D.A.), an American Association for Cancer Research/Aflac Scholar-in-Training Award (to J.R.C.), an ACS Postdoctoral Fellowship (to J.D.A.), National Institutes of Health (NIH) Transformative R01 Grant DK-085714 (to M.T.), Stand Up to Cancer Innovative Research Grant SU2C-AACR-IRG1109 (to M.T.), a Damon Runyon-Rachleff Innovation Award (to M.T.), a Prostate Cancer Foundation Creativity Award (to M.T.), Department of Defense Prostate Cancer Research Program Award W81XWH-09-1-0144 (to C.S.H.), Royalty Research Grant R21 AI095023 (to F.H.) from the University of Washington, NIH P01 Grant CA-85859 (to C.M.), and Pacific Northwest Prostate Cancer Specialized Program of Research Excellence Grant P50-CA-097186 (to C.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408301111/-/DCSupplemental.
References
- 1.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101(3):942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: Demonstration of a pathway for receptor shedding. Eur J Cell Biol. 1984;35(2):256–263. [PubMed] [Google Scholar]
- 3.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 5.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: A message in a bottle. Biochim Biophys Acta. 2012;1826(1):103–111. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14(7):14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci USA. 2013;110(43):17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huan J, et al. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73(2):918–929. doi: 10.1158/0008-5472.CAN-12-2184. [DOI] [PubMed] [Google Scholar]
- 10.Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119(3):756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32(22):2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 14.Ohno S, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran S, Palanisamy V. Horizontal transfer of RNAs: Exosomes as mediators of intercellular communication. Wiley Interdiscip Rev RNA. 2012;3(2):286–293. doi: 10.1002/wrna.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119(3):646–648. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- 17.Leslie M. Cell biology. NIH effort gambles on mysterious extracellular RNAs. Science. 2013;341(6149):947. doi: 10.1126/science.341.6149.947. [DOI] [PubMed] [Google Scholar]
- 18.Sverdlov ED. Amedeo Avogadro's cry: What is 1 μg of exosomes? BioEssays. 2012;34(10):873–875. doi: 10.1002/bies.201200045. [DOI] [PubMed] [Google Scholar]
- 19.Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine (Lond Print) 2011;7(6):780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;3(2006):3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 21.Hessvik NP, Phuyal S, Brech A, Sandvig K, Llorente A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta. 2012;1819(11-12):1154–1163. doi: 10.1016/j.bbagrm.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Bryant RJ, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata-Kawata H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajos-Michniewicz A, Duechler M, Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347(1):29–37. doi: 10.1016/j.canlet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288(48):34343–34351. doi: 10.1074/jbc.M113.480822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59(5):823–831. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014 doi: 10.3402/jec.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng HH, et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS ONE. 2013;8(7):e69239. doi: 10.1371/journal.pone.0069239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momen-Heravi F, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394(10):1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol Ther. 2012;136(2):169–174. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- 38.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 2013;59(11):1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalra H, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13(22):3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 41.Steinman RM, Brodie SE, Cohn ZA. Membrane flow during pinocytosis. A stereologic analysis. J Cell Biol. 1976;68(3):665–687. doi: 10.1083/jcb.68.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park CK, et al. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron. 2014;82(1):47–54. doi: 10.1016/j.neuron.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Reports. 2014;6(1):211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morello M, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12(22):3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delorme-Axford E, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA. 2013;110(29):12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strobl H, et al. flt3 ligand in cooperation with transforming growth factor-beta1 potentiates in vitro development of Langerhans-type dendritic cells and allows single-cell dendritic cell cluster formation under serum-free conditions. Blood. 1997;90(4):1425–1434. [PubMed] [Google Scholar]
- 49.Gatti E, et al. Large-scale culture and selective maturation of human Langerhans cells from granulocyte colony-stimulating factor-mobilized CD34+ progenitors. J Immunol. 2000;164(7):3600–3607. doi: 10.4049/jimmunol.164.7.3600. [DOI] [PubMed] [Google Scholar]
- 50.Lamparski HG, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 51.Andrews PA, Velury S, Mann SC, Howell SB. cis-Diamminedichloroplatinum(II) accumulation in sensitive and resistant human ovarian carcinoma cells. Cancer Res. 1988;48(1):68–73. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.