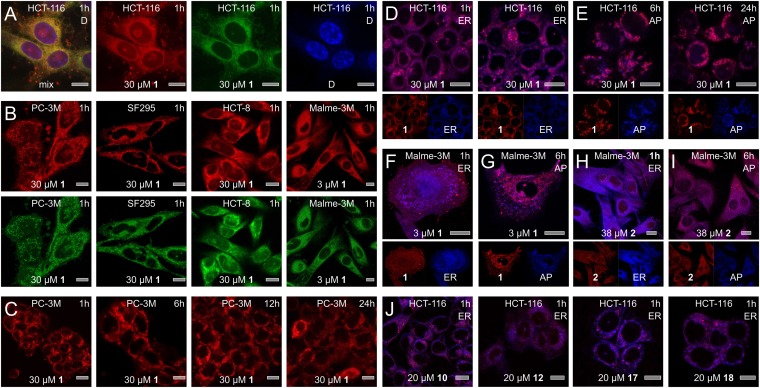
Fig. 2.
Subcellular localization studies. (A) Fluorescence from 1 was characterized by excitation maxima at 250 nm (ε = 20,800 cm−1 M−1), 289 nm (ε = 24,000 cm−1 M−1), and 342 nm (ε = 6,900 cm−1 M−1) and a broad emission from 490 to 680 nm, providing green–red fluorescence. Treatment of HCT-116 cells with 1 for 1 h results in the appearance of green–red fluorescence from 1 around the nucleus, as indicated by counterstaining with DAPI. (B) Similar subcellular localization was observed across a panel of cell lines that demonstrated sensitivity to 1. (C) Time-course analysis. Within 1 h, 1 localizes within the ER then induces autophagocytosis with 1 localizing in the forming autophagosomes (AP). (D–G) Counterstaining of the ER and AP was used to validate the subcellular localization events. Images depict staining of cells with ER stain at 1 and 6 h and AP stain at 6 and 24 h. (F–G) Similar localization was observed in Malme-3M cells albeit at lower concentrations of 1. (H–I) Treatment of Malme-3M cells with 2 also results in comparable but slower localization H, in the ER followed by I, the transition to AP. (J) Analogs 10, 12, 17, and 18 also undergo ER localization followed by transitioning into the forming AP. Each image is marked with cell line (Top Center), compound (Bottom Center), dose (Bottom Center), time (Top Right), and counterstain (Top Right). Counterstains are indicated by the following labels: ER = Tracker blue-white (ER), AP = dansylcadavarine (autophagosomes), and D = DAPI (nucleus). Bars denote 10 µm.