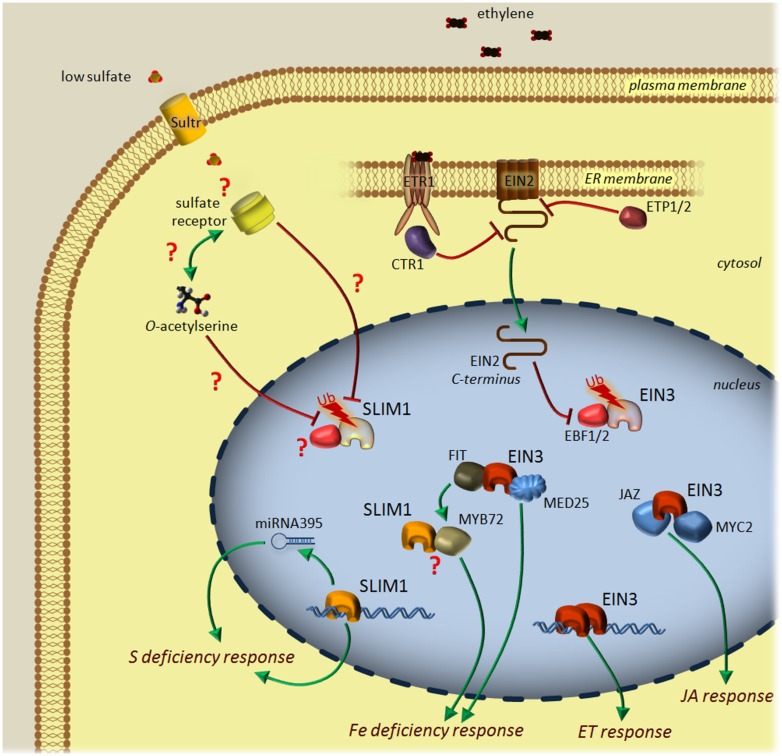
FIGURE 1.
Current model of the ethylene (ET) and sulfur deficiency signaling pathways in Arabidopsis. In contrast to ET signaling, sulfur deficiency signaling is poorly characterized. Sulfate is transported to cytosol via sulfate transporters of the Sultr family. Low sulfate availability is sensed by an unknown receptor and may depend on O-acetylserine level. The low sulfur (LSU) signal is transmitted to the nucleus and putatively stabilizes transcriptional factor SLIM1. SLIM1 induces the transcription of selected genes and miRNA395, thus reprogramming the transcriptional profile to answer the sulfur deficiency conditions. ET is perceived by the receptor proteins (for example, ETR1) present in the ER membrane. When the hormone is absent, the receptors activate a Ser/Thr kinase, CTR1, that dimerizes and suppresses the ET response by inactivating EIN2 through the phosphorylation of its C-terminal end. The EIN2 protein level is negatively regulated by the F-box proteins ETP1 and ETP2 and proteasomal degradation, while two other F-box proteins, EBF1/2 serve for the degradation of the transcription factor EIN3 in the nucleus to shut off the ET response. Upon perception of ET, ETR1 inactivates CTR1 and promotes the cleaving off of the C-terminal end of EIN2 that induces the degradation of EBF1/2 after import to the nucleus. EIN3 dimerizes and activates a transcriptional cascade of ET-responsive genes. Depending on the other environmental factors, EIN3 also interacts with JAZ proteins and transcriptional factor MYC2 to shape the jasmonic acid (JA) response. Another partner of EIN3 is MED25, which is a part of a complex regulating iron homeostasis. Additionally, EIN3 binds to FIT, a central regulator of iron deficiency response affecting the transcription level of many genes, with MYB72 among them. MYB72 can interact with SLIM1; however, the outcome of this interaction is unknown. Positive (green) and negative (red) lines represent activation and downregulation processes, respectively. SLIM1 and EIN3, shown in fading colors with Ub (ubiquitin), correspond to proteins marked for proteasome-mediated degradation. Question marks depict the points that are still waiting to be addressed by researchers.