Abstract
Background
The morbidity and mortality of cancer increase remarkably every year. It's a heavy burden for family and society. The detection of prognostic biomarkers can help to improve the theraputic effect and prolong the lifetime of patients. microRNAs have an influential role in cancer prognosis. The results of articles discussing the relationship between microRNA polymorphisms and cancer prognosis are inconsistent.
Methods
We conduct a meta-analysis of 19 publications concerning the association of four common polymorphisms, mir-146a rs2910164, mir-149 rs2292832, mir-196a2 rs11614913 and mir-499 rs3746444, with cancer prognosis. Pooled Hazard Ratios with 95% Confidence Intervals for the relationship between four genetic polymorphisms and Overall Survival, Recurrence-free Survival, Disease-free survival, recurrence are calculated. Subgroup analysis by population and type of tumor are conducted.
Results
GG genotype of mir-146a may be the protective factor for overall survival, especially in Caucasian population. C-containing genotypes of mir-196a2 act as a risk role for overall survival. The same result exists in Asian population, in Non-Small Cell Lung Cancer and digestive cancer. The patients with C allele of mir-149 have a better overall survival, especially in Non-Small Cell Lung Cancer. No significant results are obtained for mir-499 polymorphisms.
Conclusions
Genetic polymorphisms in mir-146a, mir-196a2 and mir-149 may be associated with overall survival. This effect varies with different types of cancer. Genetic polymorphism in mir-499 may have nothing to do with cancer prognosis.
Introduction
Cancer is a primary cause of morbidity and mortality in the vast marjority of regions in the world. The world estimated incidence and mortality in 2012 is 14.09 million and 8.2 million, respectively [1], [2]. According to the development trend, the new cases in 2030 will reach 22.2 million [3]. Cancer itself and medical treatment for cancer have been a heavy burden for both family and society. A constantly increased attention, these years, focuses on the disclosure of the methods that could treat patients effectively and economically. The detection of biomarkers will help to diagnose underlying patients at an early period and the identification of targeted genetic sites can promote the theraputic effect and prolong the lifetime of patients.
With the development in medical researches, it is widely recognized that the polymorphisms in microRNA genes can act as an essential role in carcinogenesis and progression. microRNAs (miRNAs) are the endogenous, small non-coding RNAs with a length of 18–25 nucleotides. The seed region of the miRNAs can recognize and complementarily combine with the 3′UTR of the specified mRNA, thus disrupting the biosynthesis. Numerous studies have detected a higher or lower level of microRNAs in patients with poor outcome than those with good outcome [4], [5]. The polymorphisms in microRNAs may alter their ability to combine with the targeted mRNA and consequently strenghen or weaken their ability to disrupt biosynthesis [6].
Once detected, microRNA have attached much attention for its multiple roles in tumorigenesis.
miR-146a shows a more extensive role in cancer. It may target to TRAF6 [7], IRAK1 [8], and thus play an influential role in the prognosis of patients suffering inflammation after surgery or chemoradiotherapy. It can also up-regulate the expression of PDGFRA [9] to enable the regeneration capacity of endothelial cells. Two peaks of miR-146a appear in 8 h and 24 h after chemoradiotherapy in the study [10]. It can influence the expression of WASF2. WASF2 is the downstream molecular that can transmit the GTPase signal to actin skeletal, thus affect the ability to migrate [11].
Others find miR-196 family can target to HOXC8 [12] and LSP1. And the polymorphism of the gene can alter the ability [6]. HOXC may influence the ability of migration and invasian of cells and the ability depends on the ratio of the expression between mir-196a and HOXC8 mRNA [12]. The high expression of LSP1 in multiple myeloma can influence the effect of a new anticancer drug, Bortezomib, on inducing cell apoptosis.
Studies determined the direct role of miR-149 in the Forkhead Box M1(FOXM1) mRNA to prevent the EMT process, which is important in proliferation of tumor [13]. Expression of mir-149 may affect the Puma maturation to prolong the lifetime of cells [14]. In gastric cancer, miR-149 can prevent the cell cycle by down-regulating ZBTB2 protein in ARF-HDM2-p53-p21 pathway [15]. miR-149 also can induce cell apotosis by down-regulating the expression of Akt1, E2F1 and b-Myb [16], [17].
The underlying biological mechanism of mir-499 in cancer is not elucidated. Some bioinformatic tools are used to explore the potential mechanism. Two breast cancer suppressors, NBN and BCL2L14, are predicted targets of hsa-miR-499 [18].
Genetic polymorphisms in mirnas may influence the cancer prognosis either by affecting the maturation [6], [19], [20] or by altering ability to combine with target mRNAs [6]. Studies showed that SNP in mir-146a can influence the expression of mature miR-146a [20], [21]. Mir-196a2 polymorphism was observed to alter the ability to combine with target [6]. Mir-149 polymorphism can affect its ability to regulate downstream targets by affecting the maturation of miR-149 [22].
Recently, the emerging role of microRNA polymorphisms in prognosis of cancer patients attracts some interest. In different types of cancers, microRNAs show to have different roles. In glioma [23], miRNAs show the risk role for deaths, while in gastric cancer [24], they may function as a protective factor for overall survival. Although in the same type of cancer, microRNA may have different functions. This may result from the small sample size in a single study. With the controversial results, we conduct this meta-analysis to evaluate the relationship between common genetic polymorphisms in four microRNAs (mir-146a rs2910164, mir-149 2292832, mir-196a2 rs11614913, mir-499 rs3746444) with cancer prognosis. To the best of our knowledge, this is the first meta-analysis concerning the four genetic polymorphisms with cancer prognosis.
Methods
Search strategy
This meta-analysis was carried out in accordance with the guidelines of the meta-analysis of the Observational Studies in Epidemiology group (MOOSE) [25]. We took a comprehensive search strategy in this study. The search strategy used the following terms variably combined by “microRNA”, “mir”, “cancer”, “carcinoma”, “tumor”, “survival”, “overall survival”, “Recurrence”, “disease-free survival”, “recurrence-free survival”, “disease-specific survival”, “prognosis” and “prognostic”. All of the avaliable database or online sources, such as PubMed, Scie, CBM, google scholar, CNKI, WanFang, were searched; After a browse of the title and abstract, the articles, including conference abstract, original articles and reviews, were screened out; The reference lists were searched as well. The last time for search on March, 2014. Only reviews published in English were evaluated.
Eligible studies included in this meta-analysis met the following criteria: (i) Discuss the role of the four microRNA polymorphisms in cancer; (ii) Investigate the overall survival outcome or other clinical variables, such as RFS, DSS, DFS and recurrence; (iii)HR and 95%CI are accessable. Articles were excluded based on any of the following criteria: (i)Duplicated articles or data; (ii) Lack of HR and 95%CI.
Data extraction
Two authors independently extracted data. If not consistent, the third author will join in to discuss. Any controvery will be solved by voting. All the data were subject to consensus. We contacted the authors of the articles for missing data by email. We extracted information including first author's name, year of publication, origin of the study population, size of the study population, type of tumor, genotyping method, the polymorphism site, method of survival analysis, HR(95%CI), and the follow-up time(months). HR values>1 were considered indicative of significant associations with poor outcome.
Statistical methods
Heterogeneity was assessed using Q statistics (P<0.05 was considered heterogeneous). Any significant heterogeneity among the studies was resolved using the random-effects model. Otherwise, the fixed-effects model was used. The I2 statistic, which measures the percentage of the total variation across studies that is due to heterogeneity rather than to chance, was also assessed. The effect of miRNA polymorphisms on survival outcome (OS) were estimated using forest plots. Stratified analysis of pooled HR and 95%CI for the relationship between polymorphisms with cancer prognosis in various population and cancers was done. Pooled HR was calculated using a fixed-effects model or random-effects model as appropriate. Pooled HR>1 indicated poor prognosis and was considered statistically significant if the 95% CI did not contain 1 [26]. Bonferroni correction was applied to control the potential false positive error. In this meta-analysis, the multiple comparision for mir-146a, mir-196a2, mir-149 and mir-499 was performed 13, 12, 9 and 9 times, respectively. The statistically significant P-value after correction for mir-146a, mir-196a2, mir-149 and mir-499 is 0.0038(0.05/13), 0.004(0.05/12), 0.0056(0.05/9) and 0.0056(0.05/9). Publication bias was evaluated using the funnel plot and Begg's test. P>0.05 was considered indicative of a lack of publication bias [27]. Sensitivity analysis was conducted by eliminating articles one by one. All analyses were performed using STATA vision 13.0. All of the P-value is two sided and a P-value less than 0.05 was considered to be statistically significant.
Results
Study Characteristics
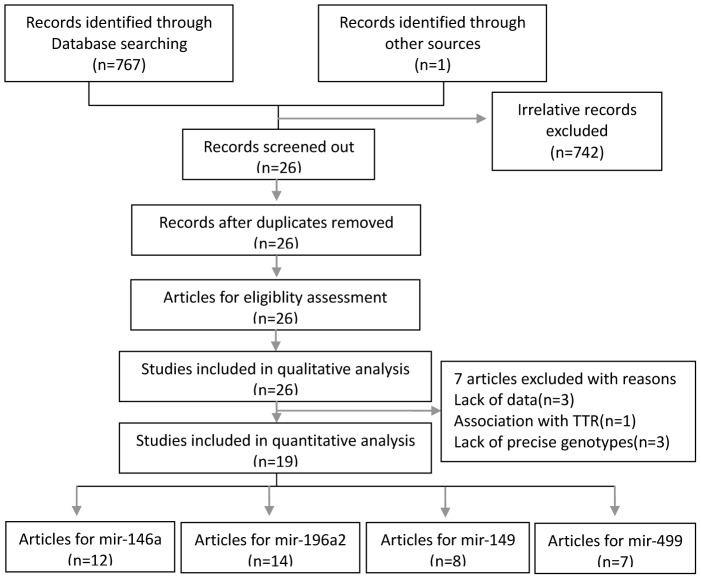
The flow diagram of the study selection process is presented in Figure 1. Nineteen [7], [23], [24], [28]–[43] eligible publications are included in this meta-analysis with 8890 patients totally. Seven [44]–[50] are excluded for lack of data and precise genotypes. These eligible articles were published from 2008 to 2014. Twelve [23], [24], [28]–[35], [37], [43] studies concerning the relationship between mir-146a polymorphism and cancer prognosis. Of them, nine articles focus on the relationship with overall survival, one on the relationship with recurrence, three on relationship with recurrence-free survival(RFS) and three on relationship with disease-free survival(DFS). The number of the articles concerning the relationship between polymorphisms in mir-196a2, mir-149 and mir-499 and cancer prognosis is respectively fourteen [7], [24], [29]–[31], [33]–[35], [37]–[41], [43], eight [24], [29], [31], [33], [34], [36], [37], [42] and seven [24], [29]–[31], [33], [34], [37]. The original population contain American, Korean, Chinese, Indian, Spainish and German. The type of tumor covers colorectal cancer (CRC), gastric cancer (GC), non-small cell lung cancer (NSCLC), esophageal squamous cell carcinoma (ESCC), hepatocellular carcinoma (HCC), bladder cancer, squamous cell carcinoma of prostate (SCCOP), head and neck squamous cell carcinoma (HNSCC), hodgkin lymphomam, nasopharyngeal and malignant lymphoma. Characteristics of eligible articles are summarized in Table S1. The original data for this meta-analysis are listed in Table S2.
Figure 1. The flowchart of the selection process.
We utilized a comprehensive searching strategy to screen out potential related articles as far as possible. 26 articles focusing on the association between the four genetic polymorphisms and cancer prognosis are screened out. 7 articles are excluded in quantitative ananlysis for lack of data to calculate pooled HR and 95%CI.
Main meta-analysis results
The meta-analysis results for relationship between polymorphisms and cancer overall survival are summarized in Table 1. The forest plot and funnel plot are listed in Figure 2 and Figure 3. The results of subgroup analysis by original population are summarized in Table 2 and the results of subgroup analysis by type of tumor are summarized in Table 3.
Table 1. Pooled HRs and 95%CIs from meta-analysis for OS.
| Snp(rs) | No. of studies | No. of patients | Model | HR(95%CI) | P-value | Heterogeneity (I2, P-value) |
| Mir-146a rs2910164 | 8 | 2906 | GG vs CC | 1.088(0.921–1.286) | 0.319 | 18.3%, 0.286 |
| 5 | 2046 | CG vs CC | 0.938(0.768–1.145) | 0.527 | 38.9%, 0.162 | |
| 5 | 1560 | DOM | 0.74(0.61–0.91) | 0.004 | 19.4%, 0.291 | |
| Mir-196a2 rs11614913 | 7 | 2577 | CC vs TT | 1.129(0.757–1.683) | 0.552 | 73.3%, 0.001 |
| 3 | 1027 | CT vs TT | 1.710(1.070–2.735) | 0.025 | 23.4%, 0.271 | |
| 7 | 2401 | DOM | 1.148(0.881–1.494) | 0.307 | 67.5%, 0.002 | |
| 6 | 1940 | REC | 1.401(1.203–1.633) | <0.001 | 42.0%, 0.111 | |
| Mir-149 rs2292832 | 6 | 2046 | CC vs TT | 0.81(0.615–1.065) | 0.131 | 37.3%, 0.172 |
| 4 | 1383 | CT vs TT | 0.748(0.585–0.955) | 0.020 | 0.0%, 0.432 | |
| 6 | 2319 | DOM | 0.747(0.638–0.875) | <0.001 | 23.6%, 0.257 | |
| 3 | 875 | REC | 0.678(0.425–1.083) | 0.104 | 36.5%, 0.207 | |
| Mir-499 rs3746444 | 5 | 2040 | GG vs AA | 0.971(0.620–1.520) | 0.897 | 0.0%, 0.771 |
| 6 | 2199 | AG vs AA | 1.025(0.866–1.214) | 0.733 | 18.8%, 0.291 | |
| 3 | 1177 | DOM | 1.104(0.787–1.549) | 0.568 | 0.0%, 0.661 |
*DOM: dominant model, REC:recessive model.
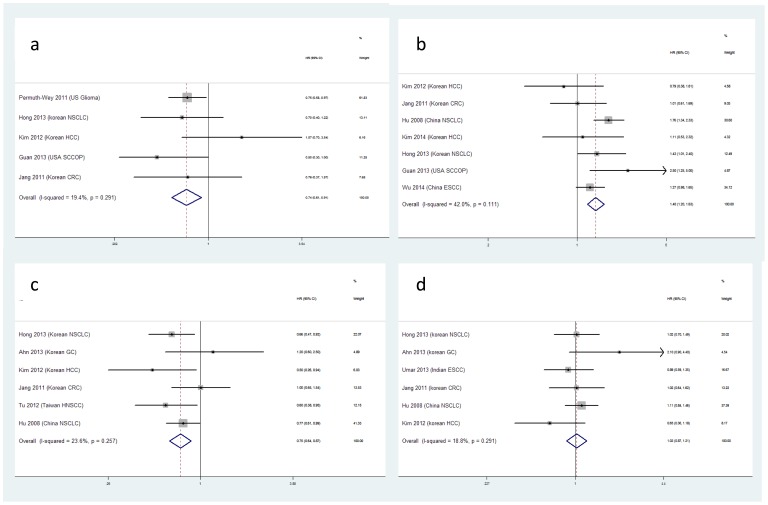
Figure 2. The main results of meta-analysis for the four genetic polymorphisms.
The forest plots for pooled HR and 95%CI estimated to demonstrate the role of mir-146a in Dominant model(a), mir-196a2 in Recessive model(b), mir-149 in Dominant model(c) and mir-499 in AG vs AA(d) in overall survival.
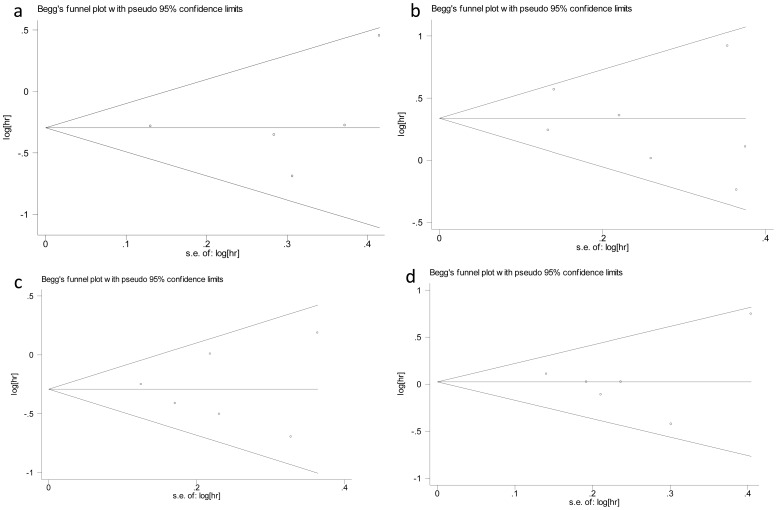
Figure 3. Funnel plots for the four genetic polymorphisms.
Funnel plots of the publication bias for mir-146a in Dominant model(a), mir-196a2 in Recessive model(b), mir-149 in Dominant model(c) and mir-499 in AG vs AA(d).
Table 2. Stratified analysis by group for different population OS.
| Snp(rs) | population | No. of studies | No. of patients | Model | HR(95%CI) | P-value |
| Mir-146a rs2910164 | Asian | 6 | 2123 | GG vs CC | 1.073(0.896–1.286) | 0.444 |
| Others** | 2 | 482 | GG vs CC | 1.179(0.768–1.810) | 0.451 | |
| Asian | 5 | 1745 | CG vs CC | 0.938(0.768,1.145) | 0.527 | |
| Asian | 3 | 922 | DOM | 0.861(0.583–1.271) | 0.451 | |
| American | 2 | 638 | DOM | 0.706(0.558–0.894) | 0.004 | |
| Mir-196a2 rs11614913 | Asian | 6 | 1917 | DOM | 1.061(0.977–1.153) | 0.161 |
| Asian | 5 | 2046 | CC vs TT | 1.086(0.901–1.310) | 0.387 | |
| Asian | 6 | 2689 | REC | 1.361(1.163–1.592) | <0.001 | |
| Mir-499 rs3746444 | Asian | 5 | 2304 | AG vs AA | 1.055(0.876–1.269) | 0.573 |
| Asian | 4 | 1887 | GG vs AA | 1.041(0.607–1.783) | 0.885 |
*DOM: dominant model, REC:recessive model.
**The others include American and Indian population.
Table 3. Stratified analysis by type of tumor for OS.
| SNP(rs) | Type of tumor | No. of study | No. of patients | Model | HR(95%CI) | P-value |
| rs2910164 | Digestive cancer | 5 | 1558 | GG vs CC | 1.116(0.897–1.388) | 0.325 |
| 3 | 1027 | CG vs CC | 0.884(0.628–1.244) | 0.479 | ||
| 3 | 895 | DOM | 0.752(0.502–1.125) | 0.166 | ||
| NSCLC | 3 | 1348 | GG vs CC | 1.051(0.812–1.361) | 0.704 | |
| 2 | 1019 | CG vs CC | 0.967(0.756–1.236) | 0.787 | ||
| rs11614913 | Digestive cancer | 5 | 1558 | CC vs TT | 0.779(0.610–0.996) | 0.046 |
| 6 | 1917 | DOM | 1.061(0.977–1.153) | 0.161 | ||
| 5 | 1670 | REC | 1.235(1.008–1.512) | <0.001 | ||
| NSCLC | 2 | 1020 | CC vs TT | 1.642(1.244–2.165) | <0.001 | |
| 2 | 1019 | REC | 1.657(1.312–2.092) | <0.001 | ||
| rs2292832 | Digestive cancer | 3 | 1027 | CC vs TT | 0.892(0.519–1.533) | 0.679 |
| 3 | 1027 | CT vs TT | 0.835(0.597–1.167) | 0.291 | ||
| 3 | 1027 | DOM | 0.875(0.636–1.204) | 0.411 | ||
| NSCLC | 2 | 1019 | CC vs TT | 0.725(0.519–1.012) | 0.058 | |
| 2 | 1019 | DOM | 0.733(0.601–0.893) | 0.002 | ||
| rs3746444 | Digestive cancer | 3 | 1021 | GG vs AA | 1.004(0.535–1.887) | 0.989 |
| 4 | 1180 | AG vs AA | 0.958(0.740–1.242) | 0.748 | ||
| NSCLC | 2 | 1019 | GG vs AA | 0.938(0.496–1.775) | 0.844 | |
| 2 | 1019 | AG vs AA | 1.078(0.862–1.347) | 0.511 |
*DOM: dominant model, REC:recessive model.
Mir-146a
In this study, we set dominant model of mir-146a as GG vs CC+CG, recessive model CC vs GG+CG. A significant result existing in dominant model indicats the protective role of homologous frequent genotype in overall survival (HR = 0.74, 95%CI 0.61–0.94, P = 0.004, Table 1). When stratified, the association between mir-146a polymorphisms and overall survival was observed in American population in dominant model (P = 0.004, Table 2). No significant association between mir-146a polymorphism and digestive cancer or NSCLC was observed in our study(Table 3). While, in Wang et al. [32] and Lin et al. studies [49], Mir-146a polymorphisms may be associated with lung cancer recurrence, moreover the polymorphisms may be related with DFS (for GG vs CC+CG, HR = 0.649, 95%CI 0.423–0.996, Table S3). We observe no association with RFS(HR = 0.669, 95%CI 0.371–1.205, Table S3).
Mir-196a2
Here, we set dominant model as CC+CT vs TT, recessive model CC vs CT+TT. CT genotype of mir-196a2 have a significantly risk role in overall survival (HR = 1.710, 95%CI 1.070–2.735, P = 0.025, Table 1). However, the association was greatly weakened after Bonferroni correction (P>0.004). Even so, a robust association was observed between CC genotype and poor overall survival in recessive model (HR = 1.401, 95%CI 1.202–1.633, P<0.001, Table 1). Consistently, the robust association was observed in Asian population (HR = 1.361, 95%CI 1.163–1.592, P<0.001, Table 2) and in digestive cancer (HR = 1.235, 95%CI 1.008–1.512, P<0.001, Table 3) and NSCLC (HR = 1.657, 95CI 1.312–2.092, P<0.001, Table 3). Moreover, C allele containing genotypes may be associated with RFS (for CT vs TT, HR = 0.675, 95%CI 0.485–0.94; for CC+CT vs TT, HR = 0.687, 95%CI 0.504–0.936, Table S3). No association with DFS (Table S3) was observed in this meta-analysis.
Mir-149
For mir-149, we set dominant model as CC+CT vs TT, recessive model CC vs CT+TT. In our study, we observe the protective role of C allele in cancer overall survival and a trend in the relationship with the number of C allele(for CC vs TT, HR = 0.81, 95%CI 0.615–1.065, P = 0.131; for CT vs TT, HR = 0.748, 95%CI 0.585–0.955, P = 0.020; for dominant model, HR = 0.747, 95%CI 0.638–0.875, P<0.001, Table 1). No significant association was observed between rs11614913 and digestive cancer overall survival in any model (Table 3). While, the genetic variant may be significantly associated with NSCLC(for CC vs TT, HR = 0.725, 95%CI 0.519–1.012, P = 0.058; for dominant model, HR = 0.733, 95%CI 0.601–0.893, P = 0.002, Table 3).
Mir-499
We set dominant model as AG+GG vs AA for mir-499 polymorphism. In our meta-analysis, we didn't gain any significant results in any model (Table 1). Results from stratified analysis indicated that mir-499 polymorphism may have no association with cancer overall survival in Asian population (Table 2). No significant association was observed between rs3746444 and digestive cancer overall survival or NSCLC in any model (Table 3).
Discussion
In this meta-analysis, we find that GG genotype of mir-146a may be a protective factor for OS, especially in Asian population. Although the statistically significant association with recurrence and DFS was detected in our study, we should notice that there are only two articles included. Nonetheless, the results imply the role of mir-146a in cancer prognosis and we should lucubrate in the future. For mir-196a2, we find an interesting matter. The C allele is a risk factor for overall survival, whereas it is a protective factor for RFS. This may result from the different types of cancers, various follow-up time period or the differences in baseline characteristics. A notable thing is that the association between mir-196a2 polymorphism and RFS is not consistent with the report in Chae [46]'s article. In Chae's article [46], a P-value larger than 0.05 is reported for the relationship between them. The article [46] is not included in this meta for it doesn't provide HR and 95%CI. This meta-analysis implies that the C allele of mir-149 may have a protective role in cancer prognosis. No statistically significant results were concluded for mir-499 polymorphisms. This may result from a relatively small number of articles discussing the association of mir-499 polymorphisms with cancer prognosis. Stratified analysis implies the association of the polymorphisms in mir-196a2 and mir-149 with NSCLC, while the association of the four polymorphisms with digestive cancer overall survival was only observed in mir-196a2 polymorphisms in this meta-analysis.
We conducted the stratified analysis by population to determine the association of these four microRNA polymorphisms with cancer prognosis. For the articles in hand, we observe that most of the studies are conducted in Asian population. Only 4, 2 and 1 are conducted respectively in Caucasion, European and Indian population. The stratified analysis by type of cancer is conducted. With a small number of articles included, the number of articles for each subgroup is 5 to the most. What a pity that we are not able to conduct stratified analysis by age, gender, somking status or other pathologic stages for insufficient articles. Some studies have reported the significant role for these polymorphisms when subgrouped by age [7], [23], [44], gender [23], [29], [44], or other pathologic stages [7], [29].
Some defects exists in our meta-analysis. Firstly, the number of articles included is relatively small, especially for mir-149 and mir-499. Secondly, we conduct stratified analysis by population, most of which are Asian, and type of tumor, most of which are digestive cancer and NSCLC, rather than other baseline characteristics. Thirdly, some heterogeneity exist in the relationship between mir-196a2 polymorphism and cancer prognosis. When exclude the Wang's article [7], the heterogeneity disapear. This may result from the different role of the polymorphism in cancer prognosis. For mir-196a2 polymorphism in Wang's article, the CC genotype is a protective factor(HR = 0.72, 0.55–0.95) in gastric cancer. Excluded, the pooled HR equals 1.476 and 95%CI ranges between 1.222–1.782 which imply the risk factor for mir-196a2 polymorphism in all cancers. This may also result from the difference in baseline characteristics. Fourthly, a P-value of 0.04 for publication bias is obtained in the association between mir-149 polymorphism and overall survival in cancers in dominant model. This may result from the small number of articles included in this meta-analysis.
Nonetheless, many advantages exist in our meta-analysis. First of all, this is the first meta-analysis concerning the relationship between the four common polymorphisms in microRNA and cancer prognosis. What's more, no heterogeneity exists in the models for the polymorphisms in mir-146a and mir-499. No publication bias is observed in the models for the polymorphisms in mir-146a, mir-149 and mir-499. Consequently, the results in our meta-analysis are stable and reliable. The last but not the least, the total number of patients in our meta-analysis is relatively large, which reaches 8057 totally.
Conclusions
All of the results observed in our meta-analysis support the role of polymorphisms in mir-146a, mir-149 and mir-196a2 in cancer prognosis, with their functions may differ from population to population, from one type of cancer to another. More studies with a larger sample size in different population are needed to determinate the role in various cancers.
Supporting Information
Basic information of the articles included in the meta-analysis.
(DOC)
The original data for the meta-analysis.
(DOC)
The association of mirna polymorphisms with DFS and RFS.
(DOC)
The seven excluded articles and the reasons.
(DOC)
PRISMA checklist.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Grant no. 81272293 and no. 81102194 from National Natural Science Foundation of China. ZBS and YZH received the funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, et al.. (2012) GLOBOCAN 2012 V1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 in.
- 2.Lyon F (2013) International Agency for Research on Cancer in.
- 3. Bray F, Jemal A, Grey N, Ferlay J, Forman D (2012) Global cancer transitions according to the Human Development Index(2008–2030): a population-based study. Lancet Oncology 13: 790–801. [DOI] [PubMed] [Google Scholar]
- 4. Huang D, Wang H, Liu R, Li H, Ge S, et al. (2014) miRNA27a Is a Biomarker for Predicting Chemosensitivity and Prognosis in Metastatic or Recurrent Gastric Cancer. Journal of Cellular Biochemistry 115: 549–556. [DOI] [PubMed] [Google Scholar]
- 5. Xia X, Yang B, Zhai X, Liu X, Shen K, et al. (2013) Prognostic role of microRNA-21 in colorectal cancer: a meta-analysis. PloS one 8: e80426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CHEN J-p, HU Z-b, TIAN T, ZHOU X-y, MIAO R-f, et al. (2009) Single nucleotide polymorphism associated with mature miR-196a2 influences the expression of Lymphocyte-specific protein 1 (LSP1) gene. ACTA UNIVERSITATIS MEDICINALIS NANJING (Natural Science) 29: 762–766. [Google Scholar]
- 7. Wang S, Tao G, Wu D, Zhu H, Gao Y, et al. (2013) A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Molecular carcinogenesis 52 Suppl 1: E87–95. [DOI] [PubMed] [Google Scholar]
- 8. Olivieri F, Lazzarini R, Babini L, Prattichizzo F, Rippo MR, et al. (2013) Anti-inflammatory effect of ubiquinol-10 on young and senescent endothelial cells via miR-146a modulation. Free radical biology & medicine 63: 410–20. [DOI] [PubMed] [Google Scholar]
- 9. Zhu K, Pan Q, Zhang X, Kong LQ, Fan J, et al. (2013) MiR-146a enhances angiogenic activity of endothelial cells in hepatocellular carcinoma by promoting PDGFRA expression. Carcinogenesis 34: 2071–9. [DOI] [PubMed] [Google Scholar]
- 10. Chaudhry MA, Omaruddin RA, Brumbaugh CD, Tariq MA, Pourmand N (2013) Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. Journal of radiation research 54: 808–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao Q, Cao Z, Tu C, Zhao Y, Liu H, et al. (2013) MicroRNA-146a acts as a metastasis suppressor in gastric cancer by targeting WASF2. Cancer letters 335: 219–24. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, et al. (2010) Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer research 70: 7894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ke Y, Zhao W, Xiong J, Cao R (2013) miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochemistry research international 2013: 506731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, et al. (2013) A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. The Journal of biological chemistry 288: 26865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, et al. (2012) MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PloS one 7: e41693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin RJ, Lin YC (2010) Yu AL (2010) miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Molecular carcinogenesis 49: 719–27. [DOI] [PubMed] [Google Scholar]
- 17. Pan S, Zhang S, Pei B, Sun Q, Bian L, et al. (2012) MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. INTERNATIONAL JOURNAL OF IMMUNOPATHOLOGY AND PHARMACOLOGY 25: 871–881. [DOI] [PubMed] [Google Scholar]
- 18. Omrani M, Hashemi M, Eskandari-Nasab E, Hasani S, Mashhadi M, et al. (2014) hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomarkers Med 8: 259–267. [DOI] [PubMed] [Google Scholar]
- 19. Lee Y, Ahn C, Han J, Choi H, Kim J, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–9. [DOI] [PubMed] [Google Scholar]
- 20. Xiong X-d, Cho M, Cai X-p, Cheng J, Jing X, et al. (2014) A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis 761: 15–20. [DOI] [PubMed] [Google Scholar]
- 21. Ramkaran P, Khan S, Phulukdaree A, Moodley D, Chuturgoon AA (2014) miR-146a Polymorphism Influences Levels of miR-146a, IRAK-1, and TRAF-6 in Young Patients with Coronary Artery Disease. Cell Biochemistry and Biophysics 68: 259–266. [DOI] [PubMed] [Google Scholar]
- 22. Ding S-L, Wang J-X, Jiao J-Q, Tu X, Wang Q, et al. (2013) A Pre-microRNA-149 (miR-149) Genetic Variation Affects miR-149 Maturation and Its Ability to Regulate the Puma Protein in Apoptosis. Journal of Biological Chemistry 288: 26865–26877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Permuth-Wey J, Thompson RC, Burton Nabors L, Olson JJ, Browning JE, et al. (2011) A functional polymorphism in the pre-miR-146a gene is associated with risk and prognosis in adult glioma. Journal of neuro-oncology 105: 639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, et al. (2013) Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Molecular carcinogenesis 52 Suppl 1: E39–51. [DOI] [PubMed] [Google Scholar]
- 25. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, et al. (2000) Meta-analysis of observational studies in epidemiology - A proposal for reporting. JAMA-J Am Med Assoc 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 26. Yusuf S, Peto R, Lewis J, Collins R, Sleight P (1985) Beta blockade during and after myocardial infarction: an overview of the randomized trials. Progress in cardiovascular diseases 27: 335–71. [DOI] [PubMed] [Google Scholar]
- 27. Sterne JAC, Egger M (2001) Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. Journal of Clinical Epidemiology 54: 1046–1055. [DOI] [PubMed] [Google Scholar]
- 28. Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, et al. (2013) A miR-146a Polymorphism (rs2910164) Predicts Risk of and Survival from Colorectal Cancer. anticancer research 33: 3233–3240. [PubMed] [Google Scholar]
- 29. Hong MJ, Choi YY, Jang JA, Jung HJ, Lee SY, et al. (2013) Association between Genetic Variants in Pre-MicroRNAs and Survival of Early-Stage NSCLC. J THORAC ONCOL 8: 703–710. [DOI] [PubMed] [Google Scholar]
- 30. Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, et al. (2013) Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Molecular carcinogenesis 52 Suppl 1: E10–8. [DOI] [PubMed] [Google Scholar]
- 31. Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, et al. (2012) Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene 504: 92–7. [DOI] [PubMed] [Google Scholar]
- 32. Wang M, Chu H, Li P, Yuan L, Fu G, et al. (2012) Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer research 72: 6173–82. [DOI] [PubMed] [Google Scholar]
- 33. Guan X, Sturgis EM, Song X, Liu Z, El-Naggar AK, et al. (2013) Pre-microRNA variants predict HPV16-positive tumors and survival in patients with squamous cell carcinoma of the oropharynx. Cancer letters 330: 233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jang MJ, Kim JW, Min KT, Jeon YJ, Oh D, et al. (2011) Prognostic significance of microRNA gene polymorphisms in patients with surgically resected colorectal cancer. Experimental and therapeutic medicine 2: 1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoon KA, Yoon H, Park S, Jang HJ, Zo JI, et al. (2012) The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancer. The Journal of thoracic and cardiovascular surgery 144: 794–807. [DOI] [PubMed] [Google Scholar]
- 36. Tu HF, Liu CJ, Chang CL, Wang PW, Kao SY, et al. (2012) The Association between Genetic Polymorphism and the Processing Efficiency of miR-149 Affects the Prognosis of Patients with Head and Neck Squamous Cell Carcinoma. PloS one 7: e51606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu Z, Chen J, Tian T, Zhou X, Gu H, et al. (2008) Genetic variants of miRNA sequences and non-small cell lung cancer survival. The Journal of clinical investigation 118: 2600–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, et al. (2010) Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 16: 3713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim HY, Yoon JH, Lee HS, Cheong JY, Cho SW, et al. (2014) MicroRNA-196A-2 polymorphisms and hepatocellular carcinoma in patients with chronic hepatitis B. Journal of medical virology 86: 446–53. [DOI] [PubMed] [Google Scholar]
- 40. Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, et al. (2013) miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Annals of surgical oncology 20 Suppl 3: S406–14. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B (2013) Single Nucleotide Polymorphisms of CASP-8 Genes, MicroRNA genes and Susceptibility and Prognosis of Malignant Lymphoma, Third Military Medical University. [Google Scholar]
- 43. Wu C, Li M, Hu C, Duan H (2014) Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum-based chemotherapy. Cancer chemotherapy and pharmacology 73: 335–41. [DOI] [PubMed] [Google Scholar]
- 44. Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, et al. (2010) Association between common genetic variants in pre-microRNAs and the clinicopathological characteristics and survival of gastric cancer patients. Experimental and therapeutic medicine 1: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, et al. (2014) Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Molecular biology reports 41: 1075–80. [DOI] [PubMed] [Google Scholar]
- 46. Chae YS, Kim JG, Kang BW, Lee SJ, Lee YJ, et al. (2013) Functional Polymorphism in the MicroRNA-367 Binding Site as a Prognostic Factor for Colonic Cancer. Anticancer Research 513–520. [PubMed] [Google Scholar]
- 47. Zheng J, Deng J, Xiao M, Yang L, Zhang L, et al. (2013) A sequence polymorphism in miR-608 predicts recurrence after radiotherapy for nasopharyngeal carcinoma. Cancer research 73: 5151–62. [DOI] [PubMed] [Google Scholar]
- 48. Xing J, Wan S, Zhou F, Qu F, Li B, et al. (2012) Genetic polymorphisms in pre-microRNA genes as prognostic markers of colorectal cancer. cancer epidemiol biomarkers prev 21: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, et al. (2010) Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 31: 1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stenholm L, Stoehlmacher-Williams J, Al-Batran SE, Heussen N, Akin S, et al. (2013) Prognostic role of microRNA polymorphisms in advanced gastric cancer: a translational study of the Arbeitsgemeinschaft Internistische Onkologie (AIO). Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 24: 2581–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basic information of the articles included in the meta-analysis.
(DOC)
The original data for the meta-analysis.
(DOC)
The association of mirna polymorphisms with DFS and RFS.
(DOC)
The seven excluded articles and the reasons.
(DOC)
PRISMA checklist.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.