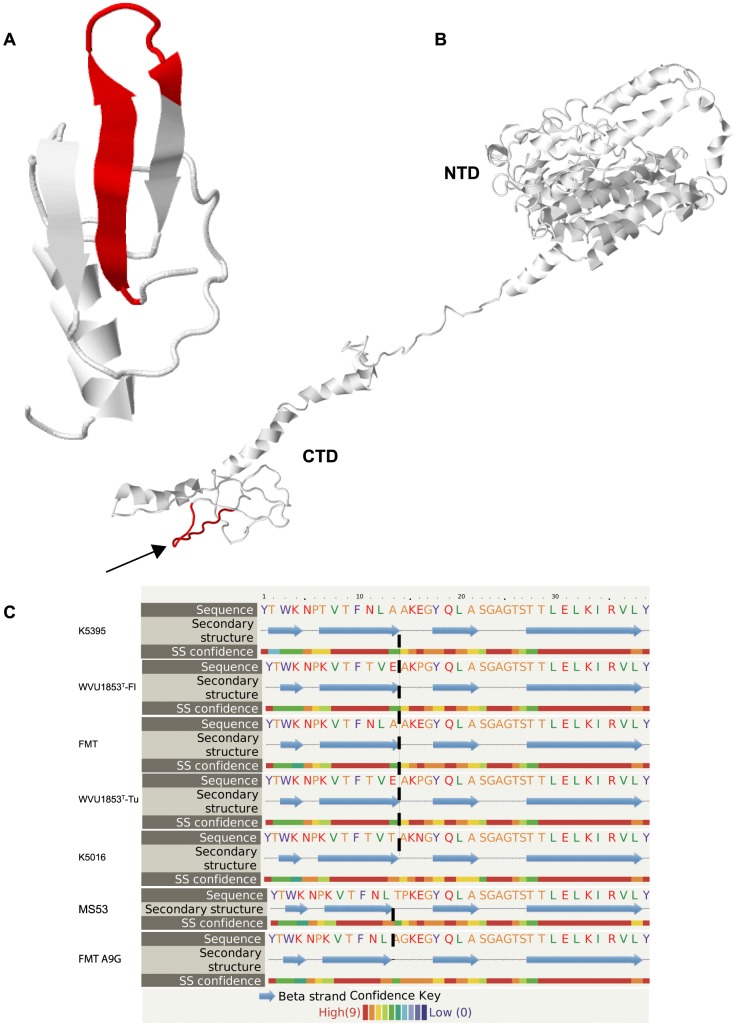
Figure 2. PHM structural predictions.
(A) The putative hemagglutination motif (PHM; red) was predicted to adopt a hairpin structure of two anti-parallel β strands separated by a short disordered loop. (B) The motif (red, indicated by arrow) mapped to a low-complexity region near the carboxyterminal domain (CTD) of the M. synoviae adhesin protein VlhA cleavage product MSPA, shown here in the structure predicted for the Tunisian lineage of strain WVU1853T. The N-terminal domain (NTD) of MSPA was predicted to have much greater 3-dimensional complexity. (C) The length of the disordered loop was predicted to be longer in PHM peptides that bound to avian erythrocyte membranes (representing Florida and Tunisian lineages of strain WVU1853T and strains FMT, K5016 and K5395) than in the reduced-binding peptide mutant FMT-Ala9Gly and the non-binding peptide representing strain MS53.