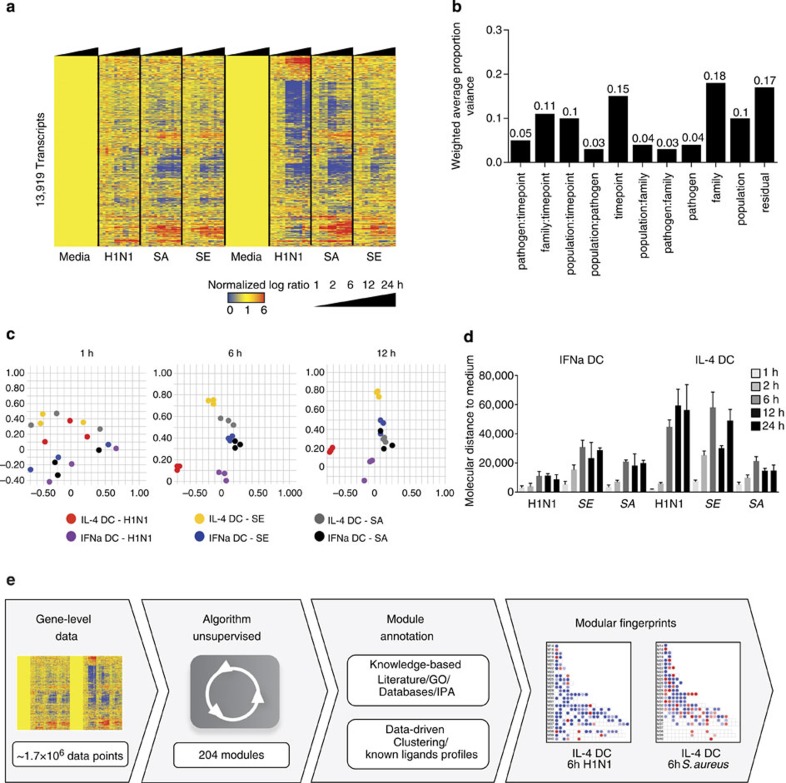
Figure 1. Transcriptional profiles of human DCs exposed to viral and bacterial pathogens over time.
(a) Heatmap of the 13,919 genes differentially expressed in DCs stimulated in vitro with influenza H1N1, S. enterica (SE) or S. aureus (SA) for 1, 2, 6, 12 or 24 h. (b) PVCA of the 13,919 genes identified in a, including DC population, pathogen, pathogen family, time point and their interacting terms. (c) Principal component analysis of the 13,919 genes differentially expressed in this data set, coloured by DC population and pathogen for 1, 6 and 12 h. (d) Bar chart representing the mean MDTM induced by the three pathogens in IL-4 and IFNα DCs at all time points as compared with medium control. Error bars represent the s.d. (three replicates). (e) Workflow describing the modular framework development process. Transcript-level data from IL-4 and IFNα DC stimulated with pathogens were automatically processed for module extraction (see Methods). Modules were annotated using a combination of knowledge-based and data-driven approaches. Finally, module fingerprints were generated for each condition studied and represented as grids or heatmaps.