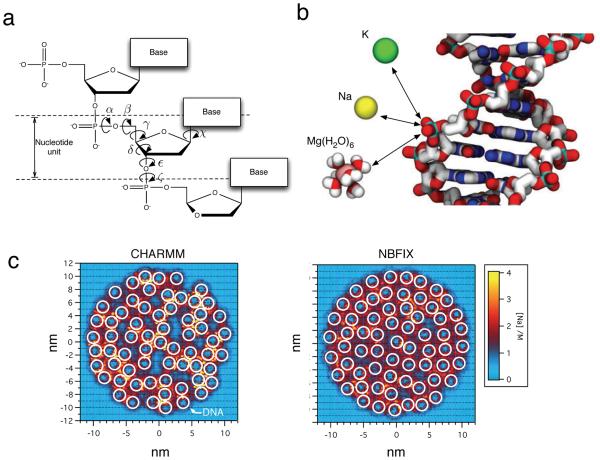
Figure 3.
Intra-DNA and DNA-ion interactions. (a) Schematic illustration of the chemical structure of DNA. Seven torsion angles are required to describe the conformation of a nucleotide unit. (b) DNA-cation interactions. The phosphate groups of the DNA backbone strongly interact with cations because of electrostatic attraction. The accuracy of these opposite-charge interactions is essential for simulations of DNA–ion and ion-mediated DNA–DNA interactions [60]. Magnesium is shown in a Mg(H2O)6 complex to emphasize its pervasive ability to coordinate water molecules. See Section 4.2.2 for a detailed discussion of Mg2+ parameterization. Bulk water and DNA hydrogen atoms are omitted for clarity. (c) Custom NBFIX corrections improve accuracy of DNA array simulations. The panels show representative configurations of 64 dsDNA molecules (white circles) confined in a cylindrical volume (not shown). The color indicates the local density of cations. Left. The artificially strong attractions between cations and DNA phosphate results in erroneous clustering of DNA. Right. Using custom parameters to describe cation–DNA phosphate interactions recovers hexagonal packing of DNA helices [60], in agreement with experiments [74].