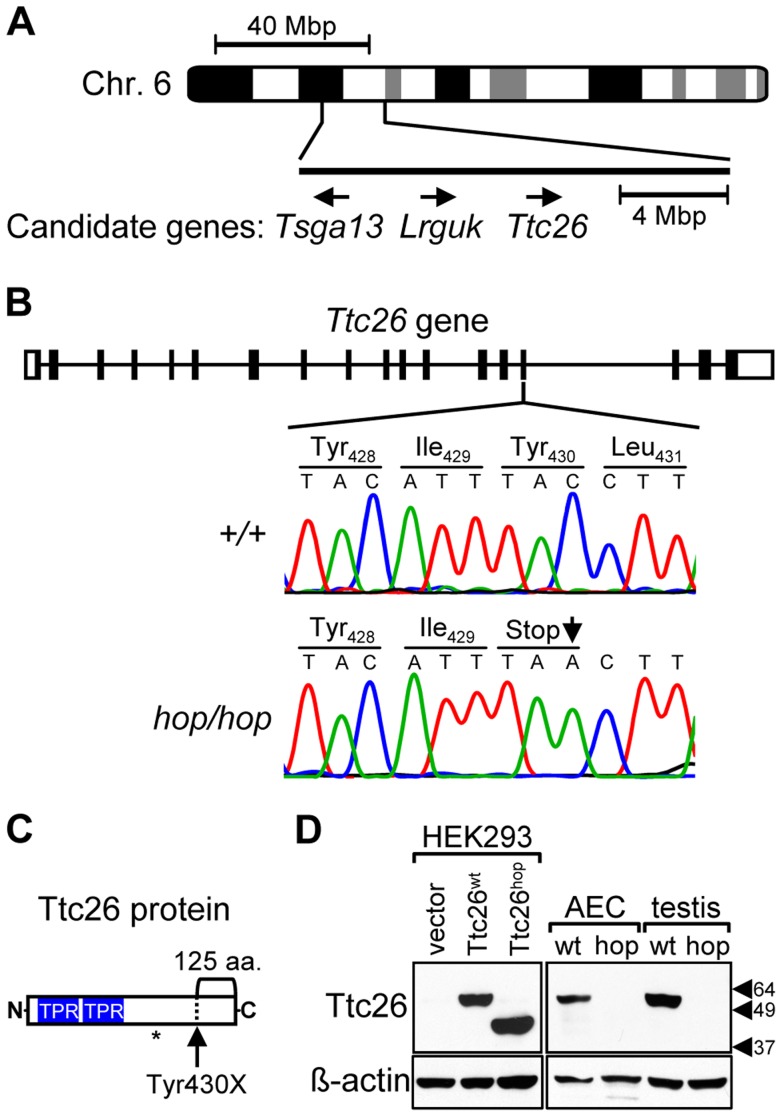
Figure 1. The Ttc26 gene of the hop mouse contains a nonsense mutation.
(A) Schematic representation of genomic positions of genes that both fall within the 16-mega base pair (Mbp) interval to which the hop mutation was mapped and had a known association with the ciliome. (B) Comparison of the 15th exon of the Ttc26 gene in wild-type and hop/hop mice. Horizontal lines represent introns, and black and white rectangles represent the coding and non-coding regions of exons, respectively. A deoxycytidine nucleotide (C) of wild-type Ttc26 (upper chromatogram) is replaced with a deoxyadenosine (A) in the hop mouse, as indicated by an arrow in the lower chromatogram. The point mutation changes the tyrosine (Tyr) at position 430 of Ttc26 to a stop codon (Stop), as shown in the amino-acid sequence lines. (C) Schematic representation of the Ttc26 protein. Blue boxes indicate the predicted TPR motifs. The bracket indicates the C-terminal 125-amino acid region of the protein that is predicted to be missing in hop/hop cells. The asterisk indicates the position of the epitope that is recognized by the anti-Ttc26 antibody. (D) Immunoblot analysis of Ttc26 expression in transfected HEK293 cells, airway epithelial cells (AEC) and testis of wild type (wt) and hop/hop (hop) mice. HEK293 cells were transfected with the indicated Ttc26-encoding construct or an empty expression vector. Arrowheads indicate the positions of the 64, 49, and 37 kDa standards. The antibodies used for immunoblotting are indicated next to the upper and lower panels.