Figure 1.
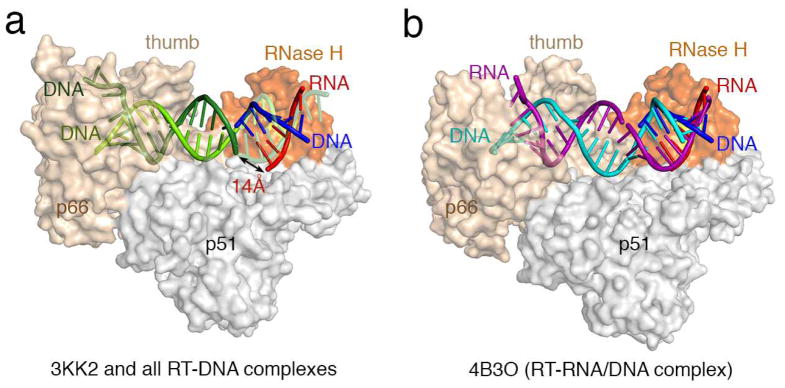
Differences between previous RT-NA complexes and our RT-RNA/DNA structures. (a) All RT-NA structrues previously reported are similar 2 and represented here by the RT-DNA-dATP ternary complex (PDB:3KK2 6) in the polymerization mode. Human RNase H1-RNA/DNA structure (PDB: 2QK9 3) is superimposed onto the RNase H domain of p66 (colored in orange) with the active site highlighted in yellow. The RNA/DNA hybrid from the human RNase H1 complex is shown in red and blue as labeled. The part of the DNA (colored light and dark green) in 3KK2 that overlaps with the RNA/DNA hybrid borrowed from the human RNase H1 complex is shown in semi-transparecy for clarity. When one DNA observed in 3KK2 (light green) and the DNA modeled into the RNase H active site (blue) are connected, the second DNA strand in 3KK2 (dark green) is 14Å from the RNA strand (red) positioned for cleavage by RNase H. (b) In all three of our RT-RNA/DNA hybrid structures, represented here by 4B3O, the RNA/DNA hybrid (purple and cyan, respectively) are A-form, where basepairs are tilted relative to the helical axis rather than perpendicular as in the B-form, and can be connected with the RNA/DNA hybrid in the RNase H active site with minor adjustment. Therefore we suggest that our RT-RNA/DNA complex structures represent the mode compatible with RNA degradation.
