Figure 2.
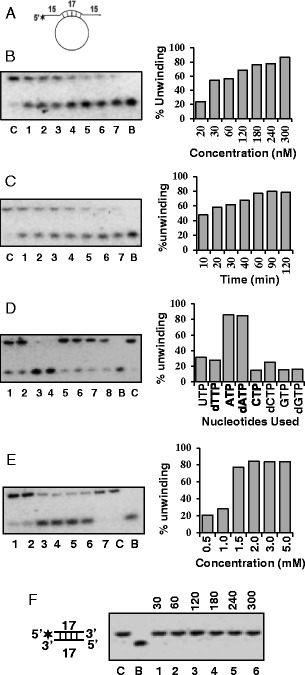
DNA helicase activity analysis of OsSUV3 protein. (A) The structure of the DNA helicase substrate used for assays in B-E. The substrate contains 15 nucleotide hanging tails at both the 5’ and 3’ ends. (B) Concentration-dependent helicase activity of purified OsSUV3, lanes 1–7 contain increasing concentration (20–300 nM) of OsSUV3. (C) Time dependent helicase activity of OsSUV3. Lanes 1–7, reactions with 250 nM of OsSUV3 at various time points. (D) Nucleotide requirement of helicase activity of OsSUV3. Lanes 1–8, helicase activity with 250 nM of OsSUV3 in the presence of various NTP or dNTPs written on the top of the autoradiogram. (E) Helicase activity with 250 nM of OsSUV3 using varying concentration of ATP. Lanes 1–6 are reactions with concentration of ATP written on the top of the autoradiogram and lane 7 is reaction without ATP. The quantitative enzyme activity data from the autoradiogram in B-E are shown as histograms on the right side. In all the B-E panels (left side), C lane is reaction without enzyme and B lane is heat denatured substrate. (F) Concentration-dependent DNA helicase activity of purified OsSUV3 protein with blunt-ended duplex DNA substrate (structure is shown in the left). Lanes 1-6 contain increasing concentration (30-300 nM) of OsSUV3 protein. Lane C is reaction without enzyme and B lane is heat denatured substrate.
