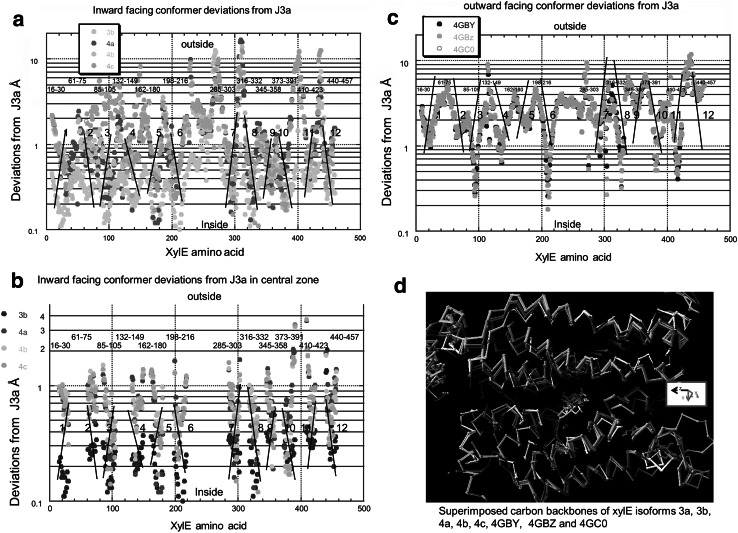
Fig. 4.
Histograms showing the RMS deviations in the Cα positions of XylE conformer 3D structures as obtained from the PDB files. These are all referenced to the inward-facing holo-conformer 3a. The same colour code is used as in Figs. 1 and 2 to signify each conformer. a shows all the RMSDs of conformers 3b, 4a,4b and 4c. b shows the RMSD of the Cα position of amino acids with the transmembrane helical regions. c shows the RMSD of the Cα positions of amino acids in outward-facing conformers 4GBY, 4GBZ and 4GC0. d Superimposed carbon backbones of XylE conformers colour coded as in Fig. 1 showing 3a, 3b, 4a, 4b, 4c, 4GBY, 4GBZ and 4GC0. The ligands binding to the high affinity central docking site are also shown. The divergence of the extramembranous strands is much more obvious than in the central region. See also Fig. S1 (Color figure online)