FIG. 2.
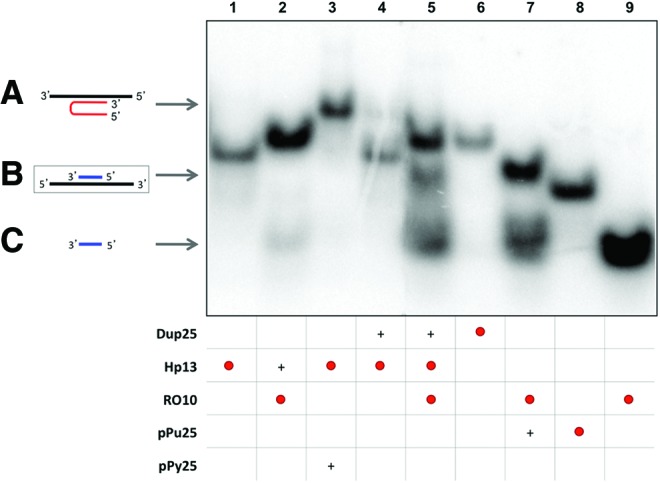
Binding between the displaced polypurine strand of the duplex (pPu25) and a specific polypyrimidine oligonucleotide (RO10). Lane 1, labeled 32P-Hp13 (2.9 nM); lane 2, specific polypyrimidine 32P-oligonucleotide (RO10) (17.1 nM) plus Hp13; lane 3, 32P-Hp13 plus polypyrimidine ssDNA (pPy25); lane 4, 32P-Hp13 plus polypurine/polypyrimidine duplex (Dup25); lane 5, 32P-Hp13 plus 32P-RO10 plus dup25; lane 6, 32P-dup25 (3.7 nM); lane 7, 32P-RO10 plus polypurine ssDNA (pPu25); lane 8, 32P-pPu25 (3.2 nM); lane 9, 32P-RO10. All unlabeled probes were used at 10 nM except for the dup25 in lane 5, which was used at 1 μM to provide an excess of duplex to be bound by the PPRH and prevent that the PPRH would sequester RO, since RO10 is also complementary to the PPRH sequence. The whole mixture of all samples was heated at 50°C before binding reaction. Incubations were performed at 37°C for 30 min. An amount of 0.5 μg of poly-dI:dC was added to each sample. Oligonucleotides are indicated by arrows. Labeled oligonucleotides are indicated as •. (+) indicate unlabelled oligonucleotides present in the reaction. (A) Triplex structure between PPRH and polypyrimidine strand (lanes 3 and 4). (B) Binding between the displaced polypurine strand and a specific polypyrimidine oligonucleotide (lanes 5 and 7). (C) Repair-oligonucleotide (RO10) (lanes 2, 5, 7, and 9). Color images available online at www.liebertpub.com/hgtb
