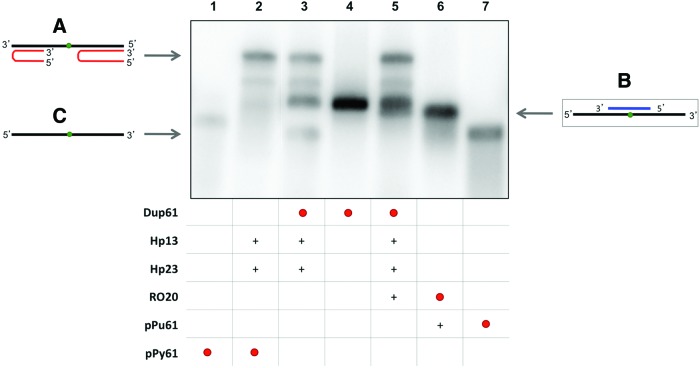
FIG. 3.
Binding between the displaced polypurine strand of the duplex (pPu61) and a specific polypyrimidine oligonucleotide (RO20). Lane 1, labeled 32P-polypyrimidine ssDNA (pPy61) (0.99 nM); lane 2, 32P-pPy61 plus hairpin-13 (Hp13) and hairpin-23 (Hp23); lane 3, 32P-polypurine/32P-polypyrimidine dsDNA (Dup61) (0.19 nM) plus Hp13 and Hp23; lane 4, 32P-Dup61 (pPu-61/pPy-61 duplex); lane 5, 32P-Dup61 plus Hp13, Hp23 and RO20; lane 6, specific 32P-RO20 (0.82 nM) plus polypurine ssDNA (pPu61); lane 7, 32P-pPu61 (0.66 nM). The entire mixture of all samples was heated at 75°C before the binding reaction. Incubations were performed at 37°C for 30 min. An amount of 1 μg of poly-dI:dC was added to each sample. Oligonucleotides are indicated by arrows. Labeled oligonucleotides are indicated as •. (+) indicate unlabelled oligonucleotides present in the reaction. (A) Triplex structure between the two PPRHs and polypyrimidine strand (lanes 2, 3, and 5). (B) Binding between displaced the polypurine strand and a specific polypyrimidine oligonucleotide (lanes 5 and 6). (C) Displaced polypurine strand (lanes 3 and 7). Color images available online at www.liebertpub.com/hgtb