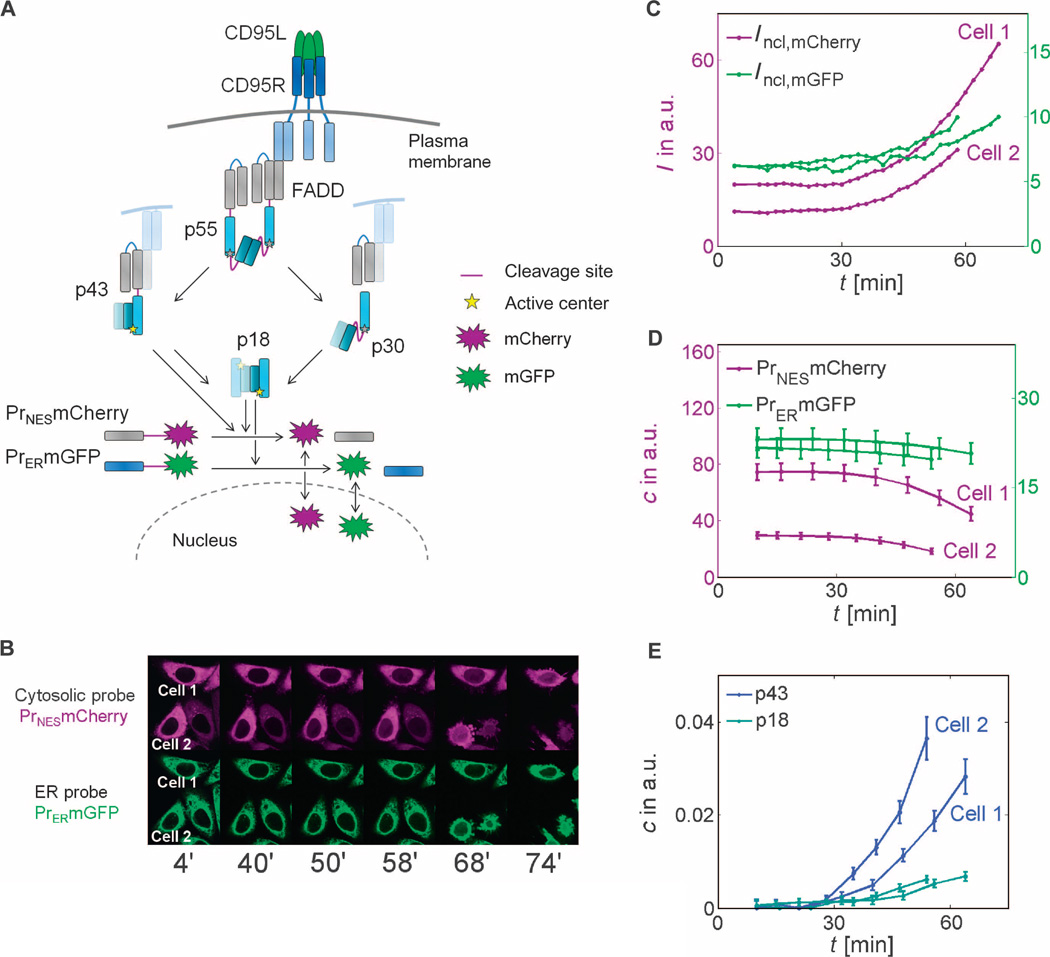
Fig. 1. Observation of single-cell caspase-8 trajectories with cleavage probes.
(A) Schematic representation of CD95 signaling. Subsequent to CD95L binding to CD95R and FADD binding, dimers of procaspase-8 (p55) are assembled and can be cut at two cleavage sites (drawn in purple). Cleavage in the catalytic domain (dark and light blue segments represent the p10 and p18 subdomains) generates p43, which is further processed in the prodomain (gray segments represent the two death domains) to p18, which enters the cytosol. Prodomain linker cleavage in p55 produces catalytically inactive p30. Whereas the cytoplasmic probe can be cleaved by p43 and p18, the ER probe is cleaved by p18 only. (B) Live-cell imaging of probe cleavage (red channel: mCherry/cytosolic probe; green channel: mGFP/ER probe). (C to E) In two marked cells, time series of nuclear fluorescence intensities (C) were measured and transformed to concentrations of uncleaved probes (D) or p43 and p18 (E) using Eqs. 2 and 3. (B to E) Quantification of enzyme activities in two exemplary CD95-HeLa cells. I, intensity; c, concentration; a.u., arbitrary units.