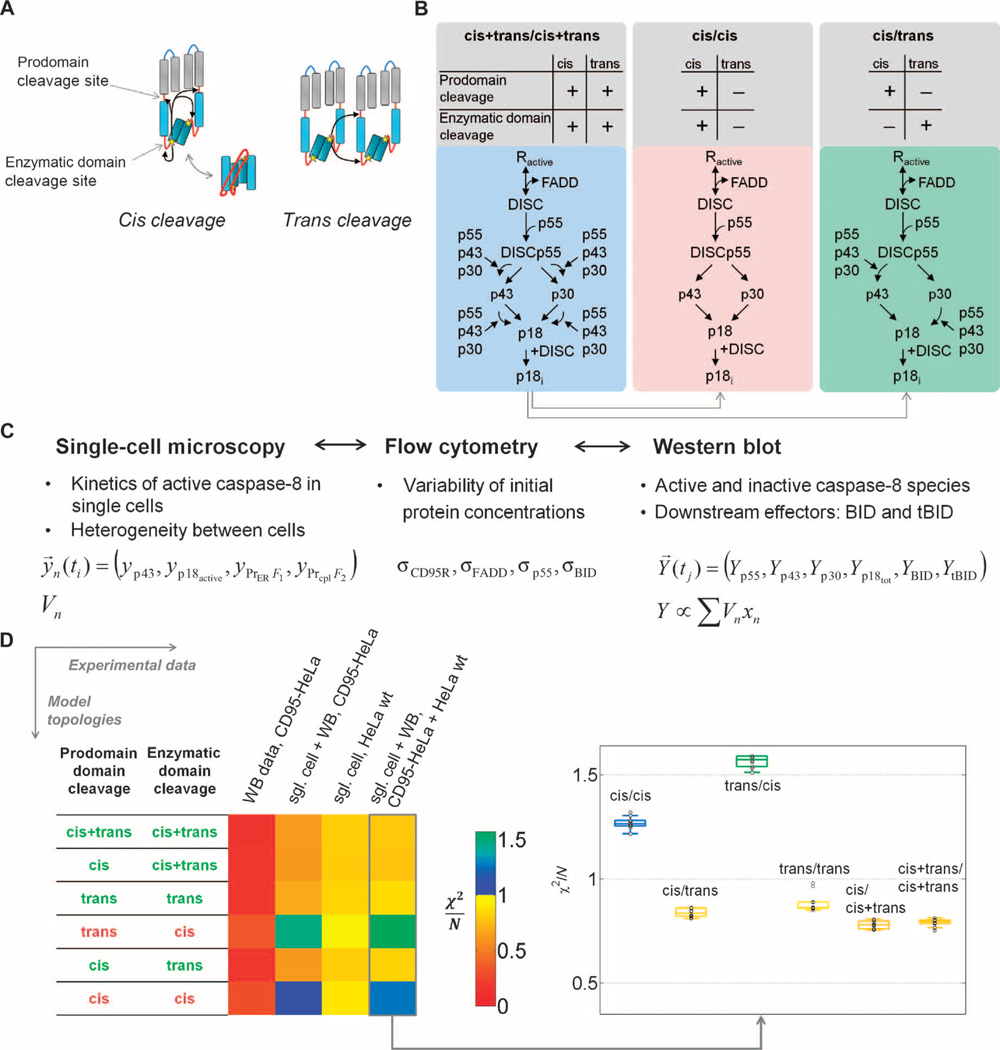
Fig. 3. Model-based discrimination of caspase-8 activation mechanisms.
(A) Intra- versus interdimeric cleavage mechanisms: Active centers (stars) in procaspase-8 can cleave domain linkers (red lines) within procaspase-8 dimers (cis) or between neighboring dimers (trans). (B) Schematic representation of model variants. The enzymatic domain sites and the prodomain sites are potentially cleaved in cis, in trans, or in both cis and trans. Here, the maximal variant cis+trans/cis+trans and two minimal variants are symbolized: a pure cis and the cis/trans topology. See fig. S6B for the other models. (C) Representation of data from different experimental techniques in the model. Single-cell models were fitted to single-cell measurements yn. Population- based measurements Y for caspase-8 intermediates, BID, and tBID are represented by sums of single-cell variables xn weighted by cellular volumes Vn. Distribution parameters from flow cytometry and mean initial protein concentrations from Western blot were not directly fitted but used to constrain parameter limits. (D) Model discrimination between cis and trans cleavage variants. Color-coded χ2/N values, indicating goodness of fit, for the 1% best of n = 1000 fits are shown for model topologies that were fit to experimental data of increasing complexity, and box plots for fits to the full experimental data set, in which box colors indicate median χ2/N values. Models with χ2/N > 1 were regarded as unable to explain experimental data.