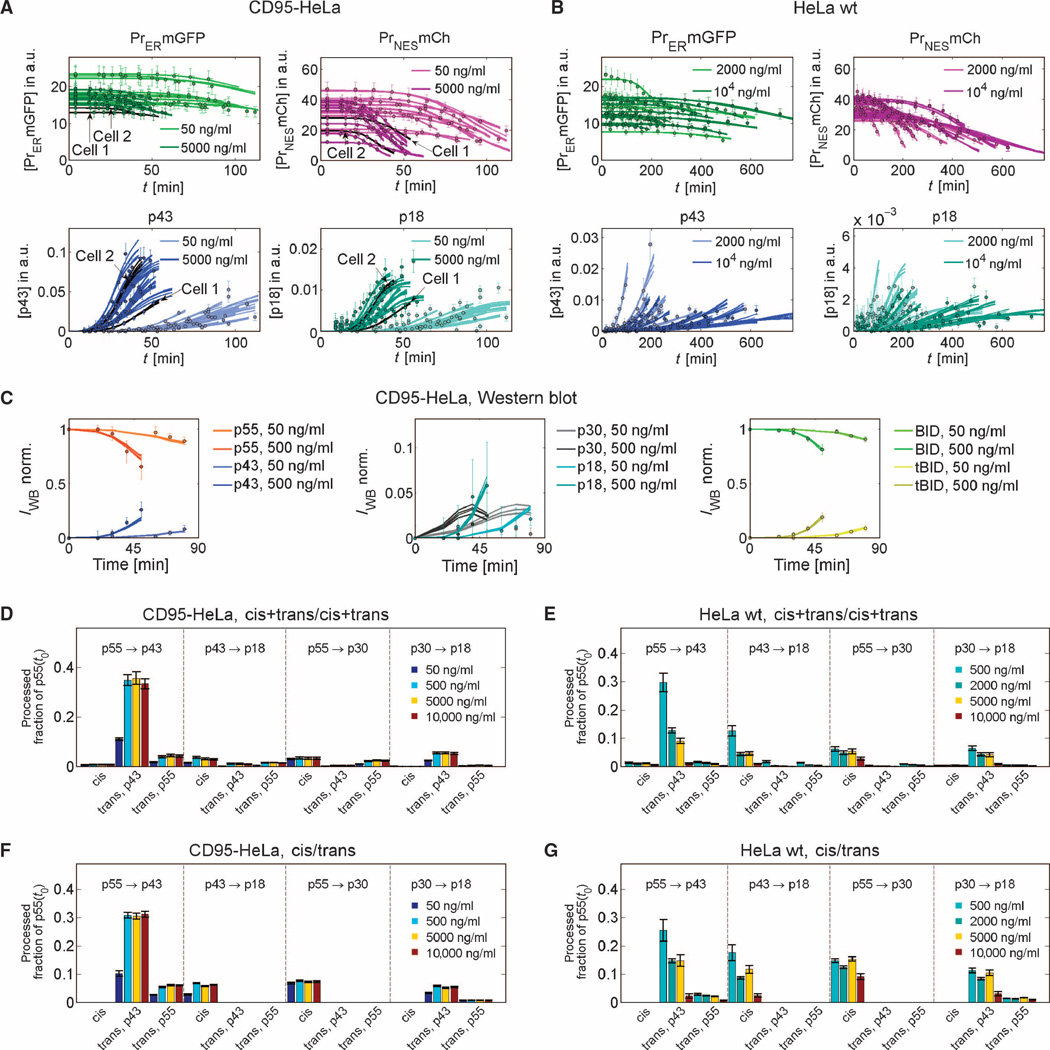
Fig. 4. Cis/trans model fits and flux analysis.
(A to C) Fits of the ensemble of single-cell cis/trans models to single-cell data of 80 cells (2 cell lines × 4 ligand concentrations × 10 cells per condition) or Western blot data (best 0.5% of n = 1000 fits). For clarity, only fits and data at two of four concentrations are shown. Experimental data (points) and model fits (lines) for cleavage probes, p43, and p18 for CD95-HeLa (A) and HeLa wt (B) cells. (C) Western blot data for CD95-HeLa cells for caspase-8 intermediates, BID, and tBID and model fits. (D to G) Flux analysis for different procaspase-8 cleavage reactions shows that the cis+trans/cis+trans model essentially behaves like the cis/trans variant. Bars indicate fractions of the initial p55 pool that are processed until median apoptosis times via different cleavage reactions at different ligand concentrations. Substrates and products of the reactions are indicated on top of the bars. For trans reactions, the enzymes are indicated below the bars. (D) and (E) show predictions of the cis+trans/cis+trans model, and (F) and (G) of the smaller cis/trans variant, for CD95-HeLa (D and F) or HeLa wt cells (E and G). Essentially, the cis+trans/cis+trans model behaves like the cis/trans version because fluxes in trans-cleavage reactions of the enzymatic domain cleavage sites (p55 to p43 and p30 to p18) and fluxes in cis-cleavage reactions of the prodomain cleavage sites (p43 to p18 and p55 to p30) are small and can be neglected. Error bars represent SDs for the best 1% of n = 1000 fits.