Figure 2.
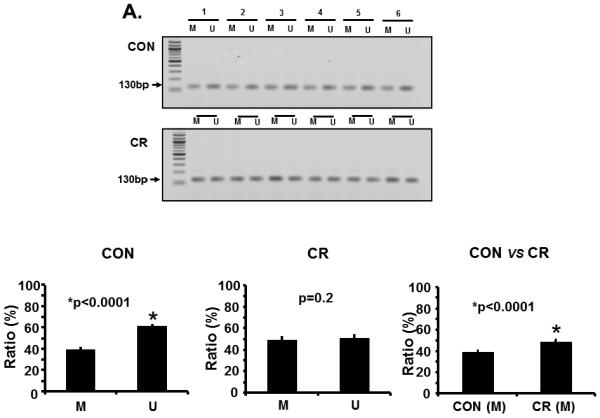
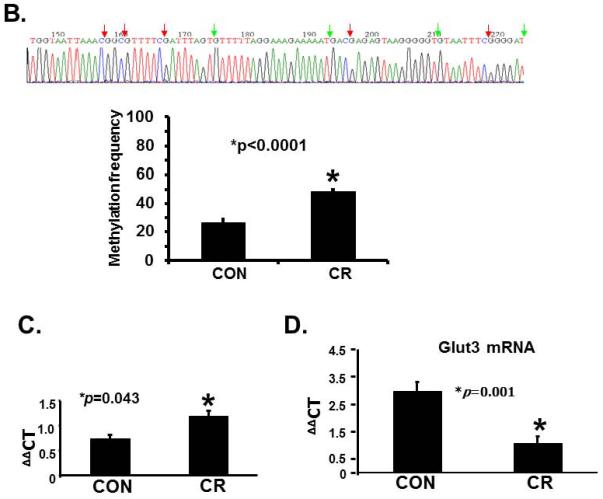
A. Representative gels demonstrating the amplification products of the glut3 5'-flanking region in control (CON; top panel) and calorie restricted (CR; bottom panel) groups by methylation specific PCR (MSP) are shown above in the inset. The numbers on top (1 to 6) represent DNA obtained from placentas of different pregnant mice. Amplified products recognized by methylated (M) and unmethylated (U) primers are seen. Relative frequency of methylation or unmethylation was calculated based on the optical density of the amplification products (arbitrary units) as a ratio of M+U value for a given sample (i.e. M=M/M=U or U=U/M+U). n=6/group, *p value as shown in each graph when compared to the corresponding CON group. The graphs shown below depict the relative frequency of methylation (M) and unmethylation (U) in CON (left) and CR (middle) groups with the methylation frequency compared between the CON and CR groups (right). B. Bisulfite sequencing of the glut3 5'-flanking region containing the CpG island. The top panel demonstrates the sequencing data with red arrows depicting the methylated and green arrows the unmethylated CpG sites within the CpG island of genomic DNA recovered from CR placentas. The bar graph in the bottom panel demonstrates the quantification of the methylation frequency in CON versus CR groups. *p value shown, n=6 clones in each experimental group. C. The bar graph demonstrates quantification of CpG island methylation by methylation specific qPCR of genomic DNA obtained from CON and CR placentas and shown as ΔΔCT values. *p value is shown, n = 6 per each experimental group. D. The bar graph demonstrates placental Glut3 mRNA concentrations assessed by reverse-transcription and real time quantitative PCR. *p value is shown. n=6 in each experimental group.
