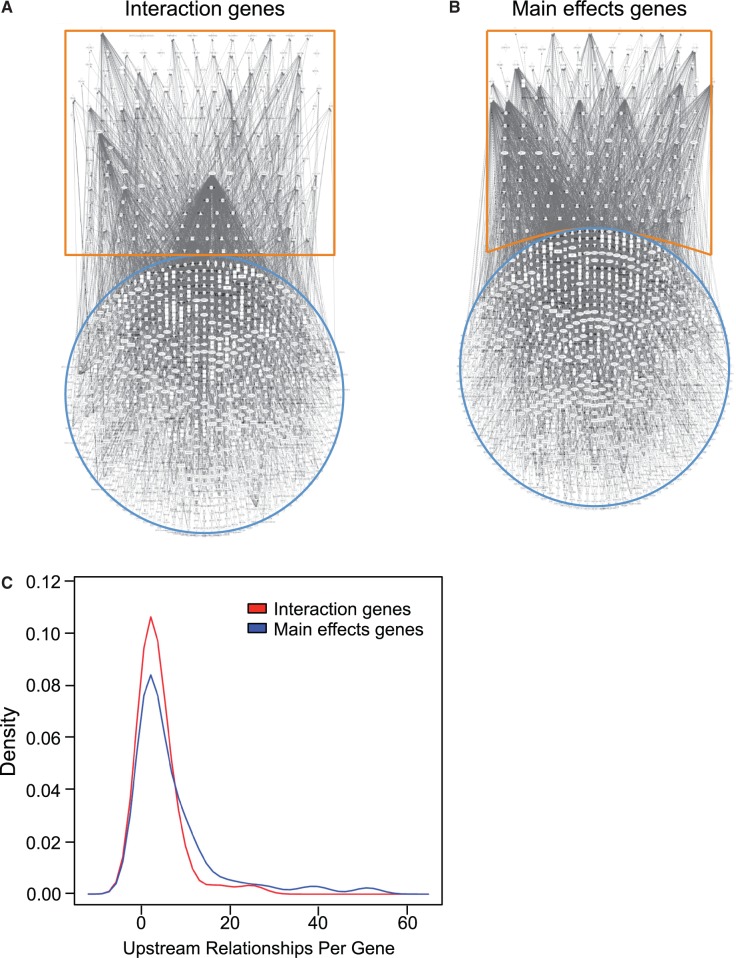
Fig. 4.
Phenotypic plasticity-enabling genes display reduced transregulatory complexity. Gene regulatory networks constructed using IPA software (Ingenuity Systems) between the uniquely interaction gene sets (A) and the uniquely main effect gene sets (B) are highlighted in orange, and their known, direct and indirect upstream transregulating molecules are highlighted in blue. Visual inspection of the networks suggests that the interaction gene sets form less complex networks (A) than noninteraction gene sets (B). Density distribution of these relationships (C) is significantly reduced in the unique interaction gene set. These networks are scale free and the probability that a vertex in the network interacts with k other vertices decays as a power law: P(k) ∼ k−g. Analysis of covariance comparing log values of P(k) to log values of k determines that the slope (g) for the interaction gene sets is significantly different (n = 13 levels of k for interaction genes, 26 levels of k for main effects genes; P < 0.05) indicating less connectivity among the genes.