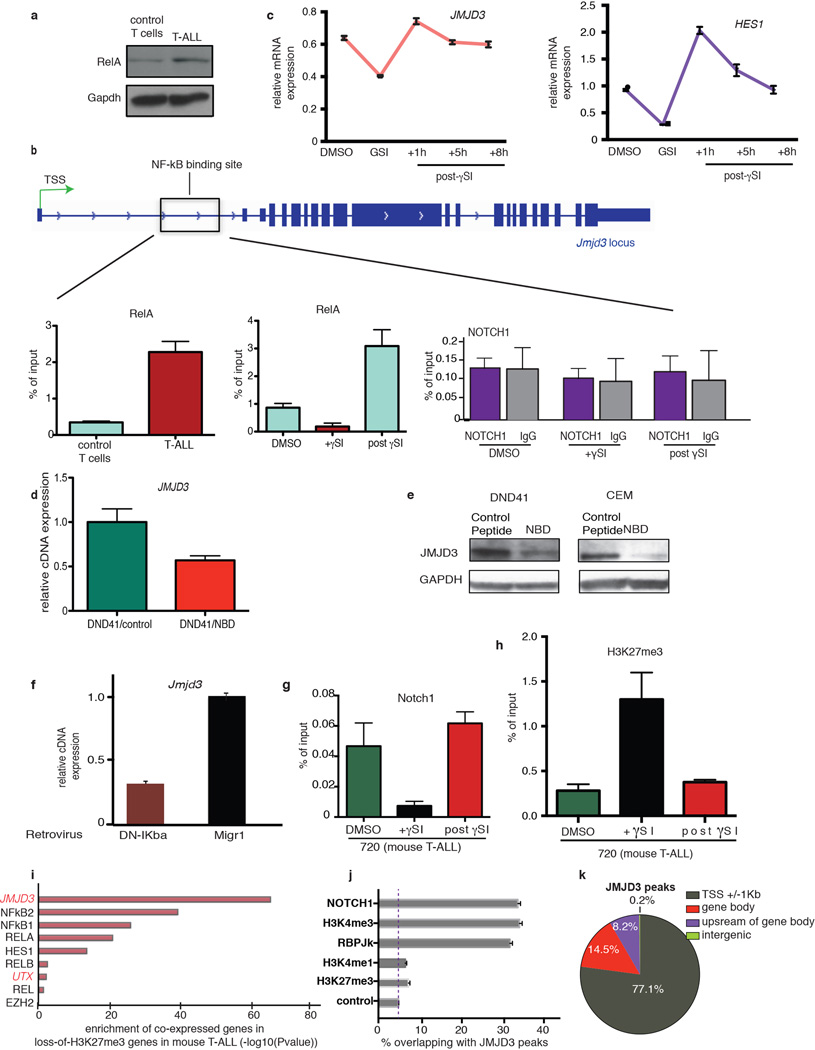
Extended Data Figure 1. MJD3 is induced through activation of the NFkB pathway in a Notch1-dependent mode in T-ALL and binds to NOTCH1 targets.
a, Levels of p65 (Rela) protein in control T cells and T-ALL tumor. A representative sample of three mice is shown. b, Schematic representation of the Jmjd3 locus showing the Rela binding site (left panel) and ChIP analysis for p65 (Rela) binding on Jmjd3 locus in mouse control T and T-ALL tumor cells as well as T-ALL cells upon treatment with gSI which affects Notch1 levels (middle panel). NOTCH1 binding to this area upon γSI treatment in T-ALL cells is also shown (right panel). c, mRNA analysis for JMJD3 and HES1 levels upon γSI treatment in CUTLL1. The average of three independent studies is shown. d, e, Expression levels of JMJD3 transcript (d) and protein (e) upon treatment of human T-ALL lines (DND41 and CEM) with NEMO binding domain (NBD) inhibitor of the NFkB pathway. f, Jmjd3 levels in T-ALL cells upon inhibition of the NFkB pathway using a dominant negative form of IkBalpha (DN-IkBa). g, h, ChIP for Notch1 (g) and H3K27me3 (h) on Hes1 promoter upon γSI treatment in murine T-ALL cells. In d and f–h the average of three studies is shown. In e a representative example of three studies is shown. i, Genes correlated with selected human genes (JMJD3, NFkB1, etc) were tested for enrichment in lossof-H3K27me3 genes during the transition to T-ALL in the mouse model. j, Overlap of JMJD3 peaks with peaks of important activating (H3K4me3 and H3K4me1) and repressive (H3K27me3) epigenetic marks as well as members of the NOTCH1 complex. The percentage of TSSs containing JMJD3 peaks is used as a conservative control, alternative to much lower genome-wide JMJD3 occupancy. k, Genome-wide distribution of JMJD3 peaks in human T-ALL.