Figure 1.
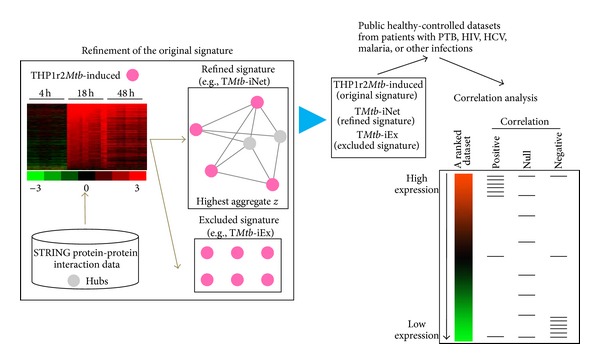
A network-based strategy to refine an expression signature. The expression signature of THP-1 macrophages in response to Mtb infection (THP1r2Mtb-induced) was used in this study [6]. To extract expression-active genes (pink dots) that are functionally linked genes, STRING protein-protein interaction data were used to select expression-active genes that interacted directly among themselves or interacted indirectly via a third protein that was defined thereby as a hub (grey dots). The degree of a hub corresponds to the number of direct interactions the hub makes. In the refined signature example, one hub has a degree of 4 and another has a degree of 3; thus the minimum degree of hubs in the example signature is 3. The selection of the minimum degree of hubs determines how transcriptionally active the refined signature will be. The final refined set was the most transcriptionally active one (i.e., TMtb-iNet, which is refined based on hubs with minimum degree 14). TMtb-iNet and the original signature (i.e., THP1r2Mtb-induced) as well as the cognate excluded genes (i.e., TMtb-Ex) were then analyzed for their correlations with other patient-derived transcriptome datasets by GSEA.
