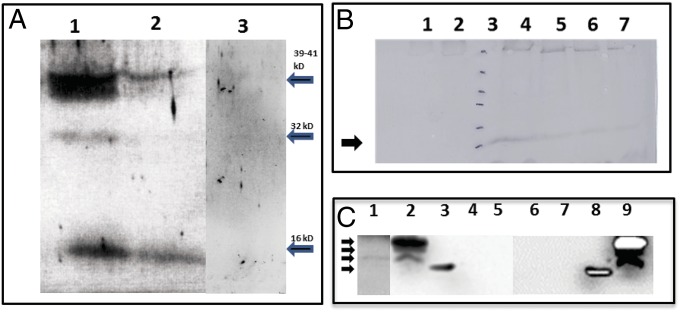
Fig. 3.
SDS/PAGE, Western detection, and time course of expression of the scRYMV-encoded proteins. (A) Western analysis using antibodies specific to the scRYMV ORF-encoded protein to detect the 16-kDa proteins among total plant proteins from RYMV infected (lane 1), healthy uninfected plant (lane 3), and purified RYMV virus (lane 2). Arrows on right indicate the positions of the 39-, 32-, and 16-kDa proteins (from top to bottom). (B) Western analysis time course of the appearance of the scRYMV 16 kDa during infection in rice plants. Lane 1 contains total proteins extracted from noninfected rice plants (negative control). Lane 2 contains total proteins from RYMV-infected plants at 4 d postinoculation. Lanes 4–7 contain total rice proteins extracted from RYMV-infected plants at 10, 14, 21, and 28 d postinoculation, respectively. Lane 3 and arrow (16 kDa) depict molecular weight size markers 116, 66, 45, 35, 25, and 18 kDa, from top to bottom, respectively. (C) North-Western analysis showing the RNA-binding activity of scRYMV-encoded proteins. Lane 1 shows the presence of the 16-kDa RNA-binding protein in infected rice, whereas lanes 2 and 9 demonstrate the detection of the 16-kDa, 18-kDa, and 19-kDa RNA-binding proteins in protein extracts from E. coli carrying pET52. Lanes 4 and 5 are the negative controls from healthy rice and pET29 protein extracts, respectively. Lanes 6 and 7 are negative control from E. coli alone and E. coli carrying pET29, respectively. Lanes 3 and 8 contain the molecular weight markers including the 14-kDa positively charged lysozyme. The 35S-labeled scRYMV and PVX probes were used in lanes 1–5 and 6–9, respectively. Arrows on left indicate the positions of molecular size markers (from top to bottom, 19, 18, 16, and 14 kDa, respectively).