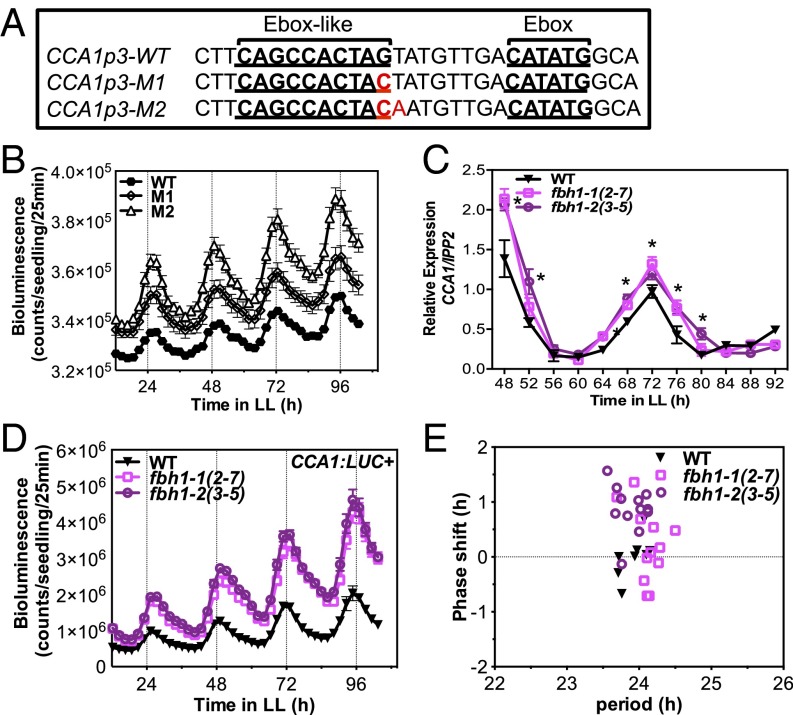
Fig. 1.
Loss of FBH1 up-regulates CCA1 expression. (A) Canonical Ebox and Ebox-like motifs present in the CCA1 promoter region −243/−42 (CCA1p3). WT and mutated versions M1 and M2 (replaced nucleotides are shown in red) were transformed into Col-0. (B) Bioluminescence analysis of CCA1p3:LUC+ expression in WT, M1, and M2 homozygous (T4) plants. (C) qRT-PCR of WT (Col-0) and FBH1 amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5). Seedlings were entrained in 12-h LD cycles for 10 d and then were released to LL. Samples were collected every 4 h for 2 d from the second full day in LL. mRNA levels were normalized to IPP2 expression. Values are shown as mean ± SD; n = 3; three independent experiments; *P ≤ 0.05; unpaired t test. (D) Bioluminescence analysis of CCA1:LUC+ expression in homozygous (F4) amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5) crossed to CCA1:LUC+ reporter lines (WT). (E) Period and phase values of luciferase expression in CCA1:LUC+ (WT) and amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5) crossed to CCA1:LUC+ lines. Period and phase values were calculated using FFT-NLLS. Only plants for which the algorithm retrieves period length and phase values are represented on the plot. In B, D, and E seedlings were entrained for 7 d in LD cycles and then imaged every 2.5 h for 5 d in LL. Values are shown as means ± SEM; n = 12.