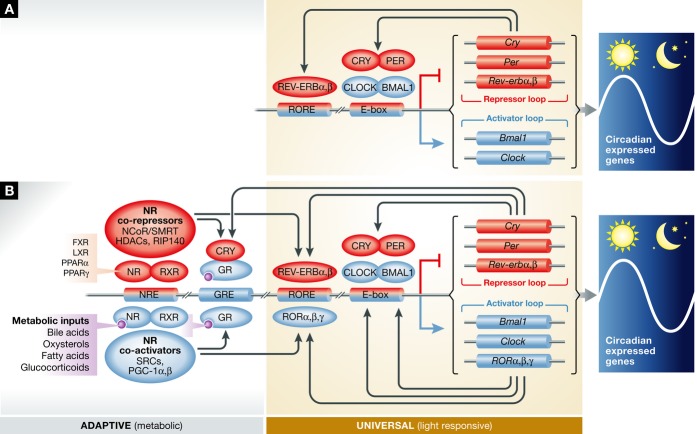
Figure 1. Canonical model of the circadian clock and the emerging model of the circadian clock in mammals.
(A) In the canonical model of the circadian clock, CLOCK and BMAL1 regulate the expression of Cry and Per. PER and CRY inhibit CLOCK and BMAL1 transcriptional activity, forming a negative feedback loop. This has long been thought to be sufficient to explain the transcriptional timing mechanism. Other factors such as REV-ERB were thought to act as a “stabilizer” 82, 83 or “output” 4. (B) In the emerging model of the molecular clock, nuclear receptors (NRs), evidenced most prominently by REV-ERBs, bind to NREs found in all core clock genes and help to orchestrate positive and negative gene expression. Many NRs are thought to share occupancy at these NREs 38, suggesting multiple orders of redundancy, compensation, and/or co-regulation at these circadian control points in the genome. Evidence of collaboration between canonical circadian clock components, such as CLOCK, PER, and CRY, arises from genome-wide ChIP-sequencing experiments, as well as classical biochemistry experiments (summarized in Table 1). Furthermore, classical NR co-regulators have been implicated in circadian time keeping.