Table 2. Kinetic equations and their parameters of the arachidonic acid/eicosanoid metabolism model.
| R1: PC → AA |
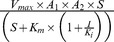
|
Vmax = 20.56 nM2s−1, Km = 2500 nM [BRENDA], Ki = 100 nM, A1 = {PLA2}, A2 = {PERK}, I = [AA] nM, S = [PC] nM |
| R2: AA → 5-HPETE |
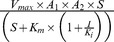
|
Vmax = 0.01 nM2s−1, Km = 0.0107 nM, Ki = 8.603 nM, A1 = {ALOX5}, A2 = {ALOX5AP}, I = [5-HPETE] nM, S = [AA] nM |
| R3: 5-HPETE → |
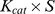
|
S = [5-HPETE], Kcat = 0.0012s−1 |
| R4: 5-HPETE → LTA4 |
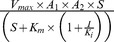
|
Vmax = 9.7953 nM2s−1, Km = 99.913 nM, Ki = 0.709 nM, A1 = {ALOX5}, A2 = {ALOX5AP}, I = [LTA4] nM, S = [5-HPETE] nM |
| R5: LTA4 → |
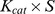
|
S = [LTA4] nM, Kcat = 0.0012 s−1 |
| R6: AA → 15-HPETE |
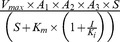
|
Vmax = 0.003 nM3s−1, Km = 0.067 nM, Ki = 1.004 nM, A1 = {ALOX15}, I = [15-HPETE] nM, A2 = {PKCD}, A3 = {PSTAT3}, S = [AA] nM |
| R7: 15-HPETE → 15-HETE |
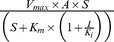
|
Vmax = 1.098 nMs−1, Km = 1.58 nM, Ki = 0.0106 nM, A1 = ALOX15, I = [15-HETE] nM, S = [15-HPETE] nM |
| R8:15-HETE → |
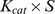
|
S = [15-HETE], Kcat = 0.00127 s−1 |
| R9: AA → PGH2 |
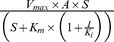
|
Vmax = 0.168 nMs−1, Km = 3.876 nM, Ki = 0.013 nM, A = {PTGS1}I = [PGH2], S = [AA] nM |
| R10: PGH2 → PGD2 |
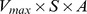
|
Vmax = 0.067 nMs−1, A = {PTGDS}, S = [PGH2] |
| R11:PGD2 → |
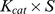
|
S = [PGD2] nM, Kcat = 0.052 s−1 |
| R12: AA → |

|
S = [AA] nM, Kcat = 0.00096 s−1 |
A, A1, A2 and A3 are the ratios of gene expression or protein levels of the respective enzymes between DDC-treated vs. control mice. S is the substrate of the corresponding reaction. Squared brackets refer to concentration values and curly braces indicate fold changes of DDC treatment vs. control. The Km value of PLA2 is taken from BRENDA [55], and other parameters of the table were fitted using the AJ mice metabolic concentrations (see results section).
