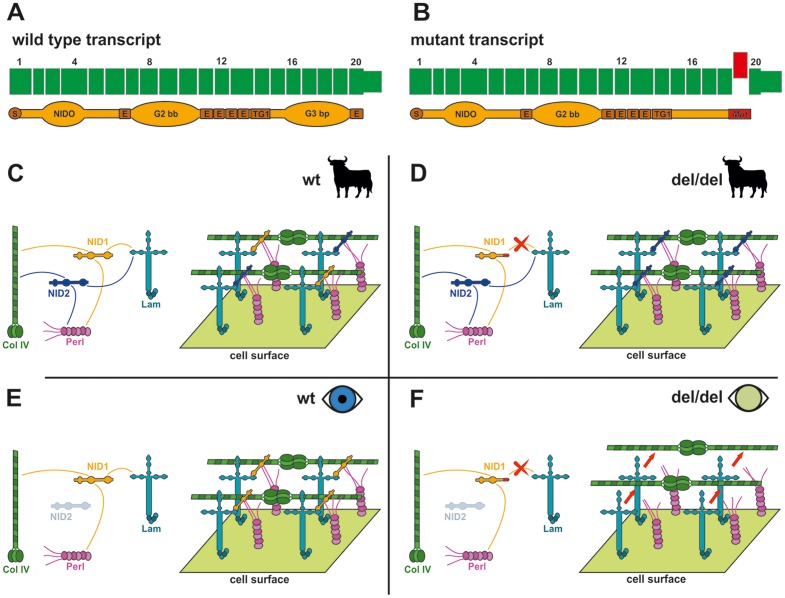
Figure 5. Tissue-specific inferred pathological mechanism.
(A) Wild type transcript. The translated NID1 protein is shown. S: Signal peptide; NIDO: Nidogen domain; E: EGF-like domains G2bb: G2 beta-barrel domain; G3bp: G3 beta-propeller domain, binding laminin [36] (B) Mutated NID1. The skipped exon is represented in red. The C-terminal part of the protein is altered severely and part of the G3 domain is lost. (C) Matrix assembly in various body tisses of an animal (like muscle, heart, kidney, skin, [31]) with wild type NID; NID1 and NID2 are both expressed in detectable level; Both interact functionally with laminin and participate to the matrix assembly, probably compensating each for the lack of the other [41]–[45]. (D) Body tissues of an animal lacking a functional NID1: in all the tissue in which NID2 is expressed (all barring brain and lenses according to literature [31]) NID2 compensates for the lack of NID1 [45]–[47]. (E) Eye of an individual with functional NID1. NID2 is not expressed at detectable levels, but NID1 alone is enough in its role of bridge protein in the matrix assembly. (F) Eye of an individual lacking of functional NID1. NID1 cannot interact functionally with laminin, and this defect is not compensated in the lens tissue, leading to matrix instability and ultimately to a pathological condition. (C-F) Simplified version of basal membrane [38]. NID1: Nidogen 1. NID2: Nidogen 2. Lam: Laminins. Col IV: Collagen IV. Perl: Perlecan. Only interactions of the Nidogens with other elements are shown. Nidogens role is to bridge the interactions among the main matrix proteins [36].