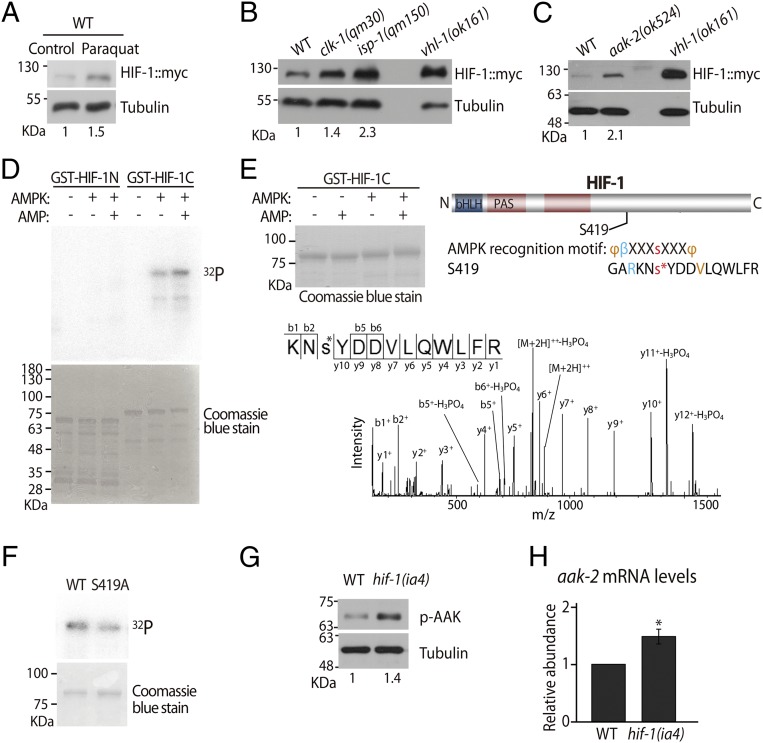
Fig. 2.
AMPK and HIF-1 are required for reciprocal down-regulation. (A and B) Western blot analysis showed that the levels of myc-tagged HIF-1 (HIF-1::myc) protein were elevated in wild-type animals by paraquat (0.25 mM) treatment (n = 5) (A) and by clk-1(qm30) or isp-1(qm150) mutations (n = 4) (B). vhl-1(ok161) mutants were used as a positive control, and α-tubulin was used as a loading control. (C) Immunoblot analysis showed that the aak-2(ok524) mutations increased HIF-1::myc protein levels (n = 5). (D) AMPK phosphorylated the C-terminal half of HIF-1 (GST-HIF-1C) in an AMP-dependent manner but not the N-terminal half of HIF-1 (GST-HIF-1N) in vitro (n = 3). Coomassie staining is shown to indicate total proteins. (E) MS analysis identified S419 as a phosphorylation site that is modestly conserved with a typical AMPK recognition motif. bHLH, basic helix–loop–helix domain; PAS, PER-ARNT-SIM domain; φ, hydrophobic residues; β, basic residues. Note the upward shift of HIF-1C by AMPK with AMP, which appears to indicate phosphorylation. MS/MS spectra recorded using a Q Exactive mass spectrometer for the doubly charged peptide KNs*YDDVLQWLFR (MH+ = 1763.81584, z = +2, XCorr = 4.55). Daughter ions are annotated according to the nomenclature for peptide fragmentation in MS. The asterisk represents the phosphorylated serine residue. (F) GST-HIF-1C S419A mutation reduced the phosphorylation level by AMPK with AMP (n = 2). Also see SI Appendix, Fig. S2 for another HIF-1 phosphorylation candidate site. We also tested the possibility that AMPK may down-regulate HIF-1 via target of rapamycin, but the data were negative (SI Appendix, Fig. S2D). (G) hif-1(ia4) mutations increased phosphorylated AAK-2 (p-AAK-2) levels (n = 5). (H) aak-2 mRNA levels were increased by hif-1 mutation as measured using qRT-PCR (n = 3). The number below each lane of the Western blot data indicates the relative band intensity of HIF-1::myc or p-AAK-2. See SI Appendix, Fig. S2 for a detailed analysis and the legend of that figure for further discussion. Error bars represent SEM; *P < 0.05, two-tailed Student's t test.