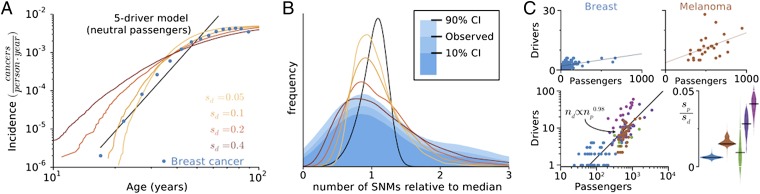
Fig. 2.
Signatures of balance between drivers and passengers in incidence and genomics data. (A) Predicted and observed breast cancer incidence rates verses age. Incidence rates in our model and the data both plateau at old age, but a traditional driver-only model (I ∝ tk) does not. (B) Histogram (blue) of the collective number of protein-coding mutations (SNMs) in breast cancer, alongside predicted distributions (lines colors as in A). Our model captures the width and asymmetry of the distribution when sd = 0.4, whereas a driver-only model predicts a narrower, less-skewed distribution. (C) (Upper) The total number of aggregate SCNA and SNM drivers verses the total number of passengers in sequenced tumors (points) fitted by linear regression. (Lower Left) Aggregated cancer genomics data plotted on log axes, with the y intercept from each subtype’s linear fit subtracted [lung cancer (green), colorectal cancer (MIN−, dark purple; MIN+, light purple)]. (Lower Right) The distribution of slope values obtained by bootstrapping 10,000 samples of each tumor type. All cancers exhibit positive slopes (P < 0.08 − 10−5), suggesting estimates of sp/sd of breast, 0.0060 ± 0.0010; melanoma, 0.016 ± 0.003; lung, 0.0094 ± 0.0093; colorectal, MIN− 0.028 ± 0.007 and MIN+ 0.041 ± 0.006.