Aurora B phosphorylation of the Polo kinase activation loop disrupts its binding to Map205 and central spindle microtubules, allowing it to be recruited to the site of cytokinesis.
Abstract
Drosophila melanogaster Polo and its human orthologue Polo-like kinase 1 fulfill essential roles during cell division. Members of the Polo-like kinase (Plk) family contain an N-terminal kinase domain (KD) and a C-terminal Polo-Box domain (PBD), which mediates protein interactions. How Plks are regulated in cytokinesis is poorly understood. Here we show that phosphorylation of Polo by Aurora B is required for cytokinesis. This phosphorylation in the activation loop of the KD promotes the dissociation of Polo from the PBD-bound microtubule-associated protein Map205, which acts as an allosteric inhibitor of Polo kinase activity. This mechanism allows the release of active Polo from microtubules of the central spindle and its recruitment to the site of cytokinesis. Failure in Polo phosphorylation results in both early and late cytokinesis defects. Importantly, the antagonistic regulation of Polo by Aurora B and Map205 in cytokinesis reveals that interdomain allosteric mechanisms can play important roles in controlling the cellular functions of Plks.
Introduction
The cell division cycle is regulated by reversible protein phosphorylation that is spatiotemporally coordinated. The conserved Polo kinase is essential for several events of mitosis and cytokinesis (Archambault and Glover, 2009; Zitouni et al., 2014). Polo-like kinase (Plk) family members are defined by an N-terminal kinase domain (KD) and a C-terminal Polo-Box domain (PBD), which mediates protein interactions (Lowery et al., 2005; Park et al., 2010). In humans, Plk1 is the closest orthologue of Drosophila melanogaster Polo in its essential roles in cell division (Petronczki et al., 2008).
The complex functions of Plks are enabled by several regulatory mechanisms (Archambault and Glover, 2009; Zitouni et al., 2014). The PBD allows Polo to interact with substrates and adaptor proteins that recruit Polo to discrete locations in the cell including centrosomes, centromeres, and the midbody (Archambault and Glover, 2009; Park et al., 2010). The PBD is a phospho-binding module, and many of its interactions are facilitated by prior phosphorylation of the partner (Elia et al., 2003a,b; Park et al., 2010). However, some PBD-dependent partners of Plks do not require phospho-priming. This is the case for Map205, a microtubule-associated protein that binds and stabilizes Polo. Instead of promoting formation of the complex, phosphorylation of Map205 at a Cdk site in early mitosis negatively regulates its interaction with Polo (Archambault et al., 2008).
Like several kinases, Plks are activated by phosphorylation in their T-loop (Qian et al., 1999; Archambault and Carmena, 2012). In humans, this phosphorylation of Plk1 occurs at Thr210 and is mediated by Aurora A kinase, with its cofactor Bora in G2 (Jang et al., 2002b; Macůrek et al., 2008; Seki et al., 2008b). Although Bora is degraded in mitosis, persisting low levels of Aurora A–Bora maintain Plk1 activity until anaphase (Chan et al., 2008; Seki et al., 2008a; Bruinsma et al., 2014). In Drosophila, we have shown that Aurora B kinase, a member of the chromosomal passenger complex (CPC), is required for T-loop phosphorylation of Polo at centromeres in early mitosis (Carmena et al., 2012a,b). Failure of the CPC to activate Polo in prometaphase leads to chromosome alignment and segregation defects (Carmena et al., 2012a).
The function of a Plk is required for cytokinesis from yeasts to humans (Petronczki et al., 2008; Archambault and Glover, 2009). In human cells, Plk1 is targeted to the central spindle via its interaction with PRC1 (Neef et al., 2007). Similarly, Drosophila Polo requires the PRC1 orthologue Fascetto for its recruitment to the spindle midzone (D’Avino et al., 2007). Plk1 is required for furrow formation by promoting the assembly of the HsCyk-4-Ect2 (RhoGAP-RhoGEF) complex, and subsequent RhoA activation and furrow ingression (Brennan et al., 2007; Burkard et al., 2007; Petronczki et al., 2007; Santamaria et al., 2007; Wolfe et al., 2009). Plk1 has been proposed to regulate additional proteins in cytokinesis, including at the midbody, before abscission (Petronczki et al., 2008; Bruinsma et al., 2012).
How Plk activities are regulated in cytokinesis is poorly understood. Here, we have investigated this question in Drosophila.
Results and discussion
Phosphorylation of Polo by Aurora B regulates its localization during cytokinesis
We showed that Polo is phosphorylated in its T-loop by Aurora B at centromeres in prometaphase (Carmena et al., 2012a). Like Polo, Aurora B localizes to the spindle midzone and midbody, and is required for cytokinesis (Ruchaud et al., 2007). Thus, we hypothesized that Aurora B could be required to activate Polo in cytokinesis.
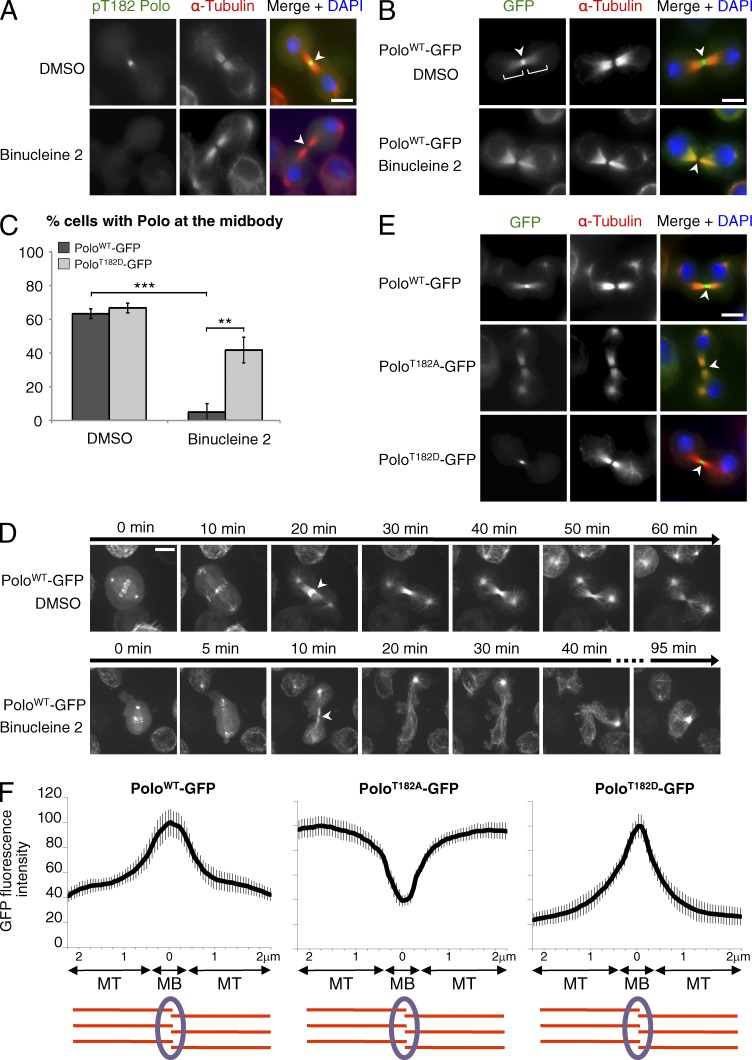
We began to test this idea using an antibody against Polo phosphorylated at its activation loop site (pT182-Polo; Carmena et al., 2012a). In Drosophila cells, pT182-Polo is detected at the midbody (Fig. 1 A). To inhibit Aurora B, we used Binucleine 2 (Smurnyy et al., 2010). Strikingly, short treatments with Binucleine 2 abrogated the pT182-Polo signal at the midbody (Fig. 1 A). To test if this result reflected the lack of phosphorylation, or a failure to recruit Polo at this site, we used stably transfected cells allowing expression of Polo-GFP at levels close to endogenous Polo levels (Fig. S1 A). Binucleine 2 treatment clearly reduced the localization of Polo-GFP at the midbody (Fig. 1, B and C; and Fig. S1 B). However, the localization of Polo-GFP to microtubules of the central spindle that are adjacent to the midbody was still visible after Aurora B inhibition. This result was confirmed by live cell imaging (Fig. 1 D and Videos 1 and 2). Moreover, Binucleine 2–treated cells failed to complete cytokinesis, as is expected upon Aurora B inhibition (Smurnyy et al., 2010; Carmena et al., 2012b). The failure of Polo to localize to the midbody was not caused by the complete absence of this structure when Aurora B was inhibited, because Binucleine 2 treatment did not disrupt the midbody localization of Aurora B, Deterin-GFP, or Pavarotti-TAP (Fig. S1, C–F). Moreover, addition of Binucleine 2 to cells with a newly formed midbody greatly shortened the retention time of Polo-GFP at that site, which indicates that Aurora B activity is required to maintain Polo at the midbody after its recruitment (Fig. S1 G).
Figure 1.
Phosphorylation of Polo by Aurora B regulates its localization in cytokinesis. (A) The localization of pT182-Polo at the midbody depends on Aurora B. Immunofluorescence in D-Mel2 cells is shown. Inhibition of Aurora B with Binucleine 2 reduced the pT182-Polo signal at the midbody (arrowheads). (B) The localization of Polo-GFP at the midbody depends on Aurora B. Binucleine 2 reduced the Polo-GFP signal at the midbody (arrowheads), but not the microtubule-associated pool of Polo-GFP (brackets). (C) Quantification of the number of cells showing clear localization of Polo-GFP or PoloT182D-GFP at the midbody after treatment with Binucleine 2 or DMSO (n = 20, repeated three times). Error bars indicate SD (**, P < 0.01; ***, P < 0.001; Student’s t test). (D) Imaging of Polo-GFP–expressing cells. Binucleine 2 or DMSO was added at anaphase onset (T0). Images were taken every 1 min. Arrowheads, midbody. (E) Mutation of Thr182 affects the localization of Polo in cytokinesis. Immunofluorescence in cells expressing PoloWT-GFP, PoloT182A-GFP, and PoloT182D-GFP is shown. (F) Line scans of the GFP fluorescence intensity along the intercellular bridge (n = 10, MT, microtubules; MB, midbody). Error bars indicate SD. Bars, 5 µm.
To test if the phosphorylation of Polo was required for its localization to the midbody, we examined the localization of a nonphosphorylatable form of Polo-GFP (PoloT182A-GFP). Strikingly, this mutant protein failed to localize to the midbody (Fig. 1, E and F). Conversely, the phosphomimetic PoloT182D-GFP mutant showed an increased localization at the midbody relative to the adjacent microtubules. Moreover, the T182D substitution largely rescued the localization of Polo-GFP at the midbody in the presence of Binucleine 2 (Fig. 1 C). Altogether, these results suggest that phosphorylation of Polo at Thr182 by Aurora B promotes Polo localization to the midbody in cytokinesis. In contrast, the T-loop state did not grossly affect the localization of Polo-GFP at kinetochores and centrosomes in early mitosis.
Phosphorylation of Polo by Aurora B promotes its dissociation from Map205
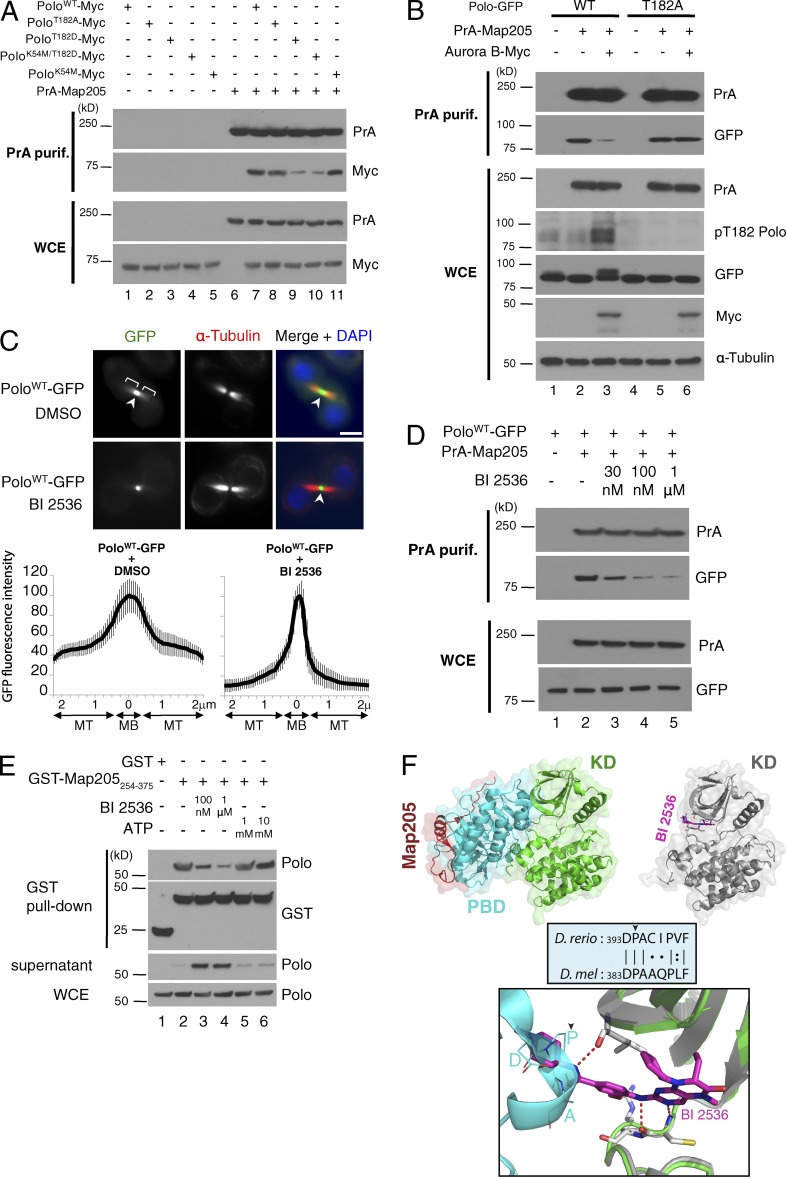
The pool of Polo that is localized on microtubules of the central spindle, and not to the midbody, is known to be bound to Map205 (Archambault et al., 2008). To investigate if T-loop phosphorylation of Polo by Aurora B could prevent or disrupt the interaction between Polo and Map205, we tested the copurification between PrA-Map205 and Thr182 mutant forms of Polo-Myc (Fig. 2 A). As expected, the association of the phosphomimetic PoloT182D-Myc mutant with PrA-Map205 was strongly diminished relative to PoloWT-Myc or PoloT182A-Myc. To test if Aurora B activity could negatively regulate the interaction between Polo and Map205, we overexpressed Aurora B–Myc in cells also expressing Polo-GFP (wild type [WT] or T182A) and PrA-Map205 (Fig. 2 B). Polo-GFP phosphorylation was increased after Aurora B–Myc overexpression, as detected by Western blotting (Fig. 2 B). This signal depended on Aurora B kinase activity because it was abolished by treatment with Binucleine 2 (Fig. S2). Interestingly, overexpression of Aurora B–Myc abrogated the copurification of PoloWT-GFP, but not PoloT182A-GFP, with PrA-Map205 (Fig. 2 B). These results strongly suggest that T-loop phosphorylation of Polo by Aurora B negatively regulates the interaction between Polo and Map205. Introduction of a kinase-dead mutation (K54M; by analogy to the K82M mutation in Plk1; Kachaner et al., 2012) in PoloT182D did not rescue its interaction with Map205 (Fig. 2 A). Therefore, the conformational change itself, induced by T-loop phosphorylation of the KD of Polo, appears to determine the affinity of the PBD for Map205.
Figure 2.
T-loop phosphorylation of Polo by Aurora B or interference with the interdomain interaction in Polo promotes its dissociation from Map205. (A) The phosphomimetic mutation T182D of Polo reduces its interaction with Map205. Cells were transfected as indicated and PrA-Map205 was purified. Purification products and whole cell extracts (WCE) were analyzed by Western blotting. (B) Overexpression of Aurora B results in hyperphosphorylation of Polo at Thr182 and abrogates its interaction with Map205. PoloWT-GFP– and PoloT182A-GFP–expressing cells were transfected with PrA-Map205 and Aurora B–Myc as indicated and treated for 1 h with okadaic acid (100 nM) to inhibit phosphatases. PrA-Map205 was purified, and Western blots were performed. (C) BI 2536 affects the localization of Polo in cytokinesis. (C, top) Immunofluorescence in cells expressing PoloWT-GFP. BI 2536 treatment (1 µM for 10 min) reduces the localization of Polo on microtubules but not at the midbody. Bars, 5 µm. (C, bottom) Line scans of the GFP fluorescence intensity along the intercellular bridge (n = 20; MT, microtubules; MB, midbody). Error bars indicate SD. (D) BI 2536 reduces the interaction between Polo-GFP and PrA-Map205. Cells were transfected as indicated and treated for 10 min with BI 2536, and PrA-Map205 was purified. Samples were analyzed by Western blotting. (E) BI 2536 disrupts the in vitro interaction between a fragment of Map205 and Polo. GST-Map205254-375 or GST-bound Sepharose beads were incubated with cell lysates and then treated with BI 2536 or ATP for 1 h as indicated. Pull-down products and the supernatant were analyzed by Western blots. (F) Binding of BI 2536 to the KD catalytic cleft is incompatible with binding of the PBD. (F, top left) Structure of the co-complex between the KD and PBD from zPlk1 and a peptide from Map205 (Protein Data Bank [PDB] accession no. 4J7B). (F, top right) Structure of human Plk1 KD bound to BI 2536 (PDB accession no. 2RKU). (F, bottom) Structural alignment of the two KD structures. BI 2536 induces a steric clash with a conserved region of the PBD comprising Pro394 involved in its intramolecular interaction with the KD. Structural alignment and rendering was performed using PyMOL 1.4 built-in commands.
The KD inhibitor BI 2536 destabilizes the Polo–Map205 complex
To test the possibility that the shift from microtubules to the midbody observed with the PoloT182D mutant was dependent on its increased kinase activity, we tested the effect of BI 2536, a widely used Plk1 ATP-competitive inhibitor, on Polo localization in cytokinesis (Steegmaier et al., 2007). Intriguingly, short treatments with BI 2536 strongly reduced the microtubules/midbody localization ratio of Polo-GFP (Fig. 2 C). Moreover, BI 2536 strongly abrogated the copurification of PoloWT-GFP with PrA-Map205 (Fig. 2 D). As the effect of BI 2536 was similar to that of the T182D activating mutation in Polo, we hypothesized that it could be caused by the induction of a conformational change, and not to kinase inhibition per se. To test this hypothesis, we devised an in vitro experiment. Recombinant GST-Map205254-375 immobilized on Sepharose was added to cell extracts to bind Polo, the resin was then washed, and the purified complex was incubated with or without BI 2536 in an ATP-free buffer. Strikingly, BI 2536 destabilized the Polo–Map205 complex (Fig. 2 E). Therefore, the effect of BI 2536 on the Polo–Map205 interaction is not caused by the inhibition of Polo kinase activity. Addition of ATP to the Polo–Map205 complex had no effect.
A cocrystal structure has recently been published between the KD of zebrafish Plk1, its PBD, and a fragment of Drosophila Map205 (Xu et al., 2013). We used structural alignment to compare the conformation of the KD in this structure to that of the KD of human Plk1 bound to BI 2536 (Kothe et al., 2007). Only subtle differences were observed between the conformations of the KDs alone. However, we noticed a steric clash between BI 2536 itself and a conserved region of the PBD including Pro394, which mediates the PBD–KD contacts in zPlk1 (Fig. 2 F; Xu et al., 2013). Thus, binding of BI 2536 in the KD catalytic cleft likely disrupts its intramolecular contact with the PBD. Together with our finding that BI 2536 induces the dissociation of Map205 from Plk1, our results suggest that the KD contact with the PBD stabilizes its Map205-interacting conformation.
T-loop phosphorylation of human Plk1 has also been reported to abrogate the KD–PBD interaction (Jang et al., 2002a). Thus, the conformational change induced by T-loop phosphorylation of Polo likely leads to the dissociation of Map205 from the PBD by weakening the PBD–KD contact (Fig. 2, A and B). Loss of the interdomain interaction in Polo would then favor the alternative conformation of the PBD, which is competent for binding to canonical phosphorylated targets (Elia et al., 2003b). Crystal structures of full-length Polo/Plk1 in their inactive and active states will be needed to fully visualize how this interdomain allosteric regulation is achieved. Activating phosphorylation of Ser137 in the PBD-contacting region of the KD of human Plk1 has also been proposed to promote the dissociation between the KD and PBD (Xu et al., 2013). However, we have never detected phosphorylation at the equivalent site (Ser109) in Drosophila Polo, and a phosphomimetic mutation of this residue did not increase Polo kinase activity (unpublished data).
Map205 sequesters unphosphorylated Polo on microtubules and inhibits its kinase activity
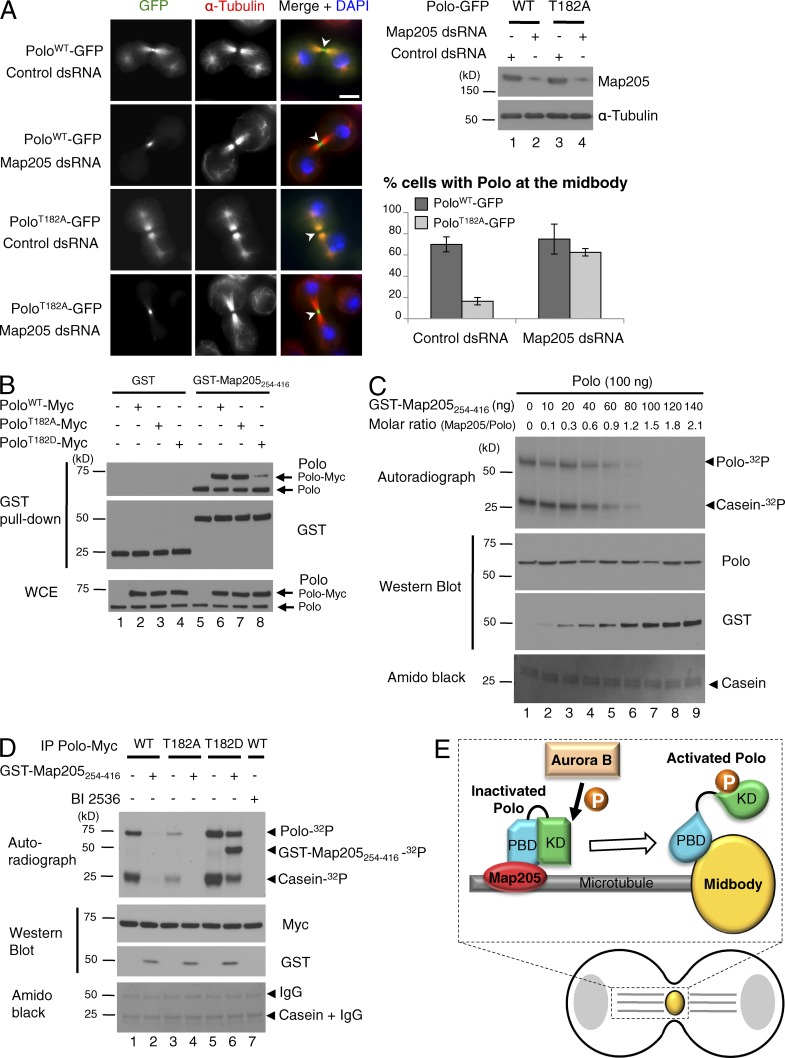
If the model we proposed is correct, then removal of Map205 should facilitate the recruitment of Polo to the midbody. As previously reported, RNAi depletion of Map205 resulted in the loss of PoloWT-GFP on microtubules but did not reduce its localization at the midbody (Fig. 3 A; Archambault et al., 2008). Strikingly, depletion of Map205 restored the localization of PoloT182A-GFP at the midbody in a majority of cells. These results confirm that when bound to Map205, Polo requires phosphorylation for its localization to the midbody because this phosphorylation induces the dissociation of Polo from Map205, and not because it promotes its kinase activation.
Figure 3.
Map205 sequesters unphosphorylated Polo on microtubules and inhibits its kinase activity. (A) Silencing of Map205 rescues the midbody localization of PoloT182A-GFP. Cells were treated for 3 d with dsRNA against Map205 or the bacterial kanamycin resistance gene (control). (A, left) Immunofluorescence showing the localization of PoloWT-GFP or PoloT182A-GFP during cytokinesis. Arrowheads, midbody. Bar, 5 µm. (A, top right) RNAi depletion of Map205 assessed by Western blots. (A, bottom right) Percentage of cytokinetic cells where PoloWT-GFP or PoloT182A-GFP were clearly localized at the midbody after RNAi treatment (n = 40, repeated three times). Error bars indicate SD. (B) A fragment of Map205 interacts with Polo in vitro. GST-Map205254-416 or GST-bound Sepharose were incubated with lysates of cells transfected with PoloWT-Myc, PoloT182A-Myc, or PoloT182D-Myc. Pull-down products were analyzed by Western blots. (C) In vitro kinase assays with Polo (100 ng) and casein as a substrate. Increasing amounts of GST-Map205254-416 were added. Reactions were analyzed by autoradiography, Western blots, and amido black (total protein). Note that Polo is capable of autophosphorylation in vitro (Fenton and Glover, 1993). (D) Kinase assays using immunoprecipitated Polo-Myc WT and mutants. Myc-tagged proteins were immunoprecipitated and used in kinase reactions in the presence of purified GST-Map205254-416 or the Polo inhibitor BI 2536 (300 nM) as indicated. (E) Model for the spatial regulation of the Polo in cytokinesis. Aurora B phosphorylates and activates Polo during cytokinesis. This event allows Polo to dissociate from Map205 and to relocalize to the midbody.
Having found that modulation of the KD could impact the Polo–Map205 interaction, we asked if, in turn, Map205 could modulate Polo kinase activity. To test it, we used a small fragment of Map205 (aa 254–416) sufficient for Polo binding (Archambault et al., 2008). GST-Map205254-416 could pull down PoloWT-Myc or PoloT182A-Myc more efficiently than PoloT182D-Myc, which is consistent with the previous results (Fig. 3 B). Strikingly, addition of GST-Map205254-416 (but not GST alone) inhibited Polo kinase activity in vitro on casein and on itself (Figs. 3 C and S3, A and B). This is consistent with the recent crystal structure between a fragment of Map205 and both domains of zebrafish Plk1. This revealed that Map205 stabilizes a closed conformation of the PBD, which in turn interacts with the KD in its inactive conformation (Xu et al., 2013). It seems reasonable to assume that Map205 inhibits Drosophila Polo enzymatic activity by the same mechanism.
To test if T-loop phosphorylation of Polo regulates its susceptibility to inhibition by Map205, we immunoprecipitated Polo-Myc WT, T182A, or T182D, and tested the effect of GST-Map205254-416 on their kinase activities (Fig. 3 D). Although PoloWT-Myc and PoloT182A-Myc could be efficiently inhibited by GST-Map205254-416, PoloT182D-Myc retained significant activity. These results suggest that Map205 preferentially binds and inhibits unphosphorylated Polo. This is consistent with the absence of pT182-Polo antibody signal on microtubules, where Polo is bound to Map205 (Fig. S1 B).
Overall, these findings suggest an interesting molecular model for Polo regulation during cytokinesis where (1) Map205 sequesters Polo on central spindle microtubules, (2) phosphorylation of Polo by Aurora B abrogates the Map205-Polo interaction, and (3), when freed from Map205, Polo is recruited to the midbody (Fig. 3 E).
Activating phosphorylation of Polo is required for cytokinesis
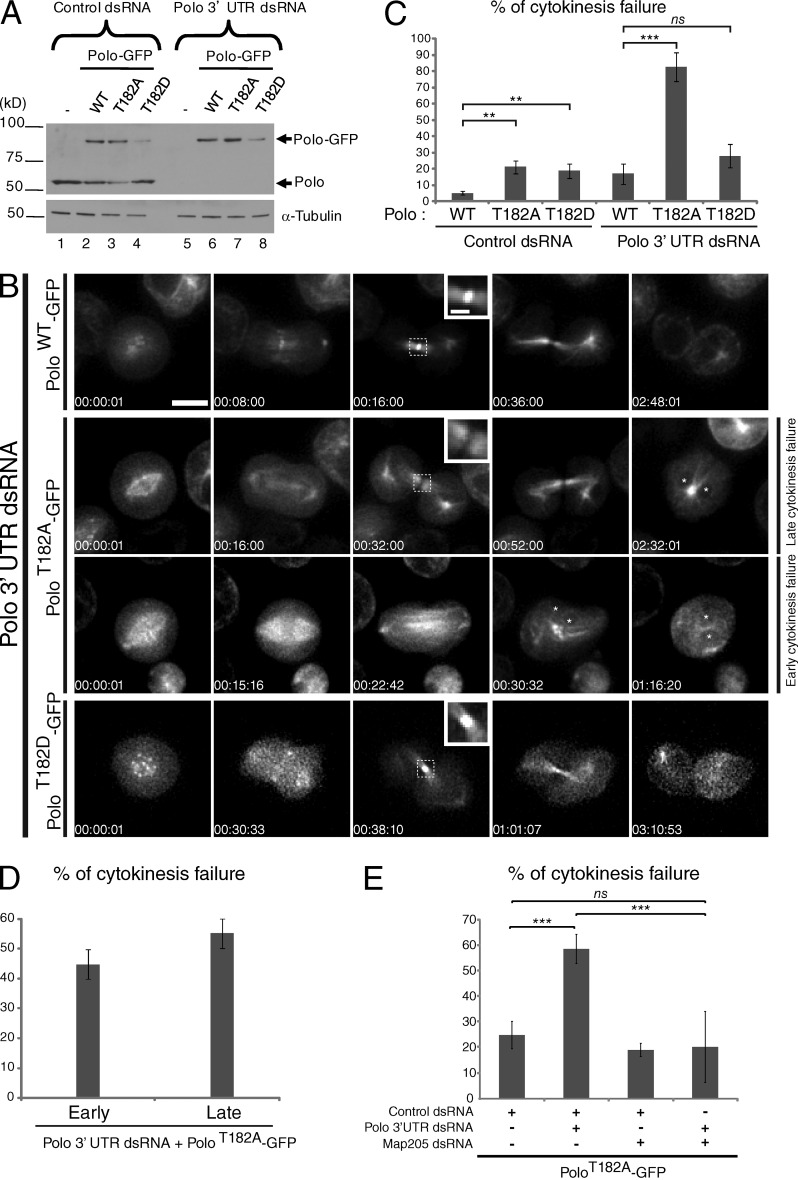
Although Polo is known to be required for cytokinesis and Polo’s full activation is known to require T-loop phosphorylation, whether Polo phosphorylation is required for cytokinesis had never been tested. Unphosphorylated Polo shows a basal kinase activity (Fig. 3 D), which we have previously shown to be sufficient for bipolar spindle assembly (Carmena et al., 2012a). We performed live imaging of cells where endogenous Polo was replaced with Polo-GFP WT, T182A, or T182D expressed at near-endogenous levels from an inducible promoter (Figs. 4 A and S1 A). Depletion of Polo in control cells by transfection of a dsRNA targeting the 3′ UTR of the endogenous transcript resulted in the accumulation of defective mitotic cells and cell death (Carmena et al., 2012a). Expression of PoloWT-GFP largely rescued cell viability and the ability of these cells to divide properly (n = 276; Fig. 4, B and C; and Video 3). Expression of PoloT182A-GFP failed to rescue the viability of Polo-depleted cells, and many cells developed early mitotic defects (Carmena et al., 2012a). Nevertheless, many cells entered anaphase and attempted cytokinesis. Of them, >80% failed cytokinesis (n = 309; Fig. 4, B–D). This failure could occur either at the onset of cytokinesis, with incomplete or no furrow ingression (45% of these cells; n = 115; Video 4), or later, as cells failed to form a stable intercellular bridge (55% of the cells; n = 142; Video 5). In both cases, failure in cytokinesis resulted in binucleation. Replacement of endogenous Polo with PoloT182D-GFP, similar to PoloWT-GFP, allowed cytokinesis to proceed in most cells (Fig. 4, B and C; and Video 6). These results indicate that T-loop phosphorylation of Polo is required for cytokinesis.
Figure 4.
Activating phosphorylation of Polo is required for cytokinesis. (A) Expression of PoloWT-GFP, PoloT182A-GFP, and PoloT182D-GFP was induced with CuSO4 (300 µM), and cells were transfected with Polo 3′ UTR dsRNA (or control KAN dsRNA). The next day, protein extracts were analyzed by Western blots. (B) Time-lapse imaging of cells depleted of endogenous Polo and expressing PoloWT-GFP, PoloT182A-GFP, or PoloT182D-GFP. Note the early or late cytokinesis failures in PoloT182A-GFP–expressing cells. Asterisks indicate nuclei in binucleate cells. Insets show enlarged views of the midbody (taken from the boxed regions). Bars: (main panels) 5 µm; (insets) 1 µm. (C and D) Quantification of cells showing a cytokinesis failure from the experiment in B. (E) Depletion of Map205 rescues cytokinesis in PoloT182A-GFP expressing cells. Cells were transfected with the indicated dsRNA and analyzed by time-lapse imaging. At least 180 cell divisions were scored for each condition in four independent experiments. Error bars indicate SD (**, P < 0.01; ***, P < 0.001; Student’s t test; ns, nonsignificant).
Polo phosphorylation could be required in cytokinesis for two reasons: (1) to activate Polo’s intrinsic kinase activity and/or (2) to free Polo from Map205 and allow its localization to the midbody. We have shown that Map205 depletion restored PoloT182A-GFP localization at the midbody (Fig. 3 A). To try to discriminate between the two possibilities, we tested if the RNAi depletion of Map205 could rescue cytokinesis in PoloT182A-GFP–expressing cells depleted of endogenous Polo. As a result, cytokinesis was largely rescued (Fig. 4 E). Although this is a complicated experiment, which should be interpreted cautiously, the result suggests that the basal activity of unphosphorylatable Polo is sufficient for cytokinesis if it can be recruited to the midbody.
Antagonistic regulation of Polo by Aurora B and Map205 in cytokinesis
Here, we showed that Aurora B and Map205 control Polo activity during cytokinesis. We had previously shown that Map205 sequesters and stabilizes Polo on microtubules in interphase, and that overexpression of Map205 in embryos interfered with Polo function (Archambault et al., 2008). In addition, Map205 inhibits Polo enzymatic activity (Fig. 3; Xu et al., 2013). This effect is the opposite of that of phosphorylated peptides derived from canonical PBD targets, which increase Plk1 kinase activity (Elia et al., 2003b). Our findings also indicate that Aurora B–dependent phosphorylation on Thr182 releases a fraction of Polo from its sequestration by Map205, and allows its recruitment to the midbody. Aurora B localizes to the midzone in anaphase and to the midbody, but it does not colocalize with the microtubule-bound pool of Polo. However, a gradient of Aurora B activity is established around the midzone in anaphase in human cells (Fuller et al., 2008). Diffusion of Aurora B from the midzone/midbody could allow it to reach and phosphorylate Polo on microtubules to promote its recruitment to the midzone/midbody. Phosphorylation of Polo by Aurora B in early mitosis likely also contributes to its dissociation from Map205, a mechanism that would collaborate with the phosphorylation of Map205 by Cdk1 (Archambault et al., 2008; Archambault and Carmena, 2012; Carmena et al., 2012a). As map205 is not an essential gene (Pereira et al., 1992; Archambault et al., 2008), control of Polo by Map205 is not essential, but our work indicates that the inhibition of Polo by Map205 imposes a need for a mechanism that allows Polo to dissociate from Map205. Indeed, the Aurora B–dependent release of Polo from Map205 is required for cytokinesis.
The requirement for Plk1 activation during cytokinesis in human cells has never been investigated. Further work will be required to determine whether human Aurora A, Aurora B, or another kinase is acting upstream of Plk1 to activate it in cytokinesis. The absence of a clear Map205 orthologue and the absence of a strong localization of Plk1 to microtubules suggest that the mechanism reported here is not strictly conserved. However, the fact that Plk1 is competent for interaction with, and inhibition by, Drosophila Map205 suggests that physiological interaction partners may regulate Plk1 in a similar way (Fig. S3 C; Archambault et al., 2008).
Importantly, our findings demonstrate for the first time that a Plk (Polo) is subject to regulation by reciprocal interdomain mechanisms for its cellular functions (Fig. 5). Our results reveal this mechanism at work in the context of a physiological process: cytokinesis. While Map205 interaction with the PBD of Polo induces inhibition of its KD, phosphorylation of the activation loop in its KD induces dissociation of Map205 from the PBD, and facilitates PBD interactions with other targets, including at the midbody. The effects of various PBD-dependent interactors on Polo kinase activity and the impact of Polo KD activation on these interactions should be examined in the context of other functions in cell division. A good candidate as an additional allosteric inhibitor of Polo is Matrimony, a PBD-dependent interactor that genetically behaves as an antagonist of Polo during female meiosis (Xiang et al., 2007). Moreover, the importance of interdomain regulatory mechanisms for the functions of other Plk family members should be evaluated. A more precise understanding of how Plk1 intramolecular switching is regulated by interaction partners, kinases, phosphatases, and small molecules could inform the development of alternative or pathway-specific Plk1 inhibitors.
Figure 5.
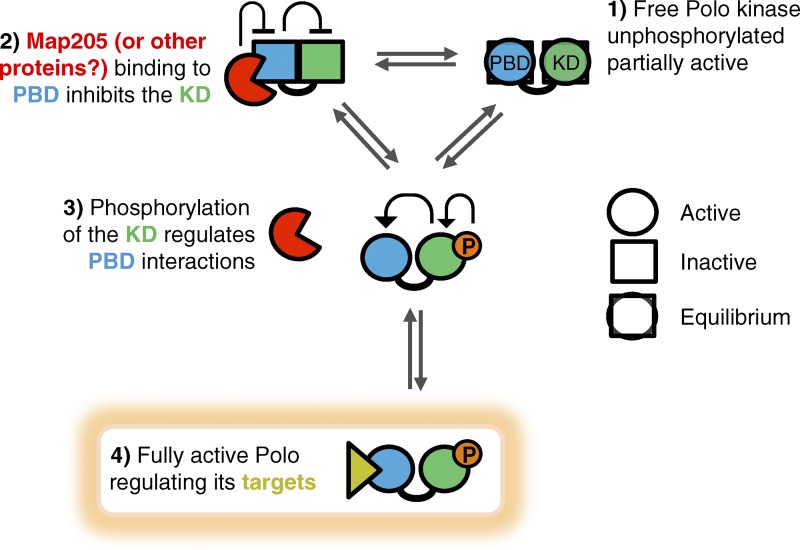
Model for the interdomain allosteric regulation of Polo. (1) Free, unphosphorylated Polo is partially active. (2) Binding of the inhibitor Map205 to the PBD stabilizes its closed conformation, which stabilizes the inactive conformation of the KD. (3) Conversely, T-loop phosphorylation of the KD induces a conformation change that destabilizes its interaction with the PBD, thereby destabilizing its Map205-binding conformation. (4) The PBD can then adopt its conformation, which is competent for binding canonical targets.
Materials and methods
DNA constructs
The expression vectors were generated by Gateway recombination (Invitrogen). Coding sequences were first cloned into the pDONOR221 entry vector. They were then recombined into the relevant destination vectors for expression from copper-inducible (pMT) or constitutive (pAC5) promoters. The following expression plasmids were generated: pMT-PoloWT-GFP, pMT-PoloT182A-GFP, pMT-PoloT182D-GFP, pAC5-PoloWT-Myc, pAC5-PoloT182A-Myc, pAC5-PoloT182D-Myc, pAC5-PrA-Map205, pAC5-Aurora B-Myc, pAC5-Deterin-GFP, and pAC5-Pavarotti-TAP. The GST-Map205254-416 plasmid has been described previously (Archambault et al., 2008), and the GST-Map205254-375 plasmid was constructed similarly, by cloning the relevant piece of Map205 cDNA into the pGEX-4T vector.
Drosophila cell culture and drug treatments
All cells were in the D-Mel2 background and were cultured in Express Five medium (Invitrogen) supplemented with glutamine, penicillin, and streptomycin. Stable cell lines expressing pAC5-Deterin-GFP, pAC5-Pavarotti-TAP, and pAC5-Aurora B-GFP were obtained by cotransfection with the pCoBlast and selection in medium containing 20 µg/ml blasticidin as described previously (Archambault et al., 2008). Stable cell lines expressing pMT-PoloWT-GFP, pMT-PoloT182A-GFP, and pMT-PoloT182D-GFP were obtained by a similar protocol but without using the pCoBlast because the Blasticidin resistance gene was inserted directly in the expression vectors. The latter cell lines were then sorted by FACS for the desired expression level of the GFP fusion proteins. Stable cell lines allowing the copper-inducible expression of Polo-GFP (WT, T182A, and T182D) were induced with CuS04 (300 µM) for at least 1 d. For Aurora B inhibition, cells were treated with either DMSO or 20 µM Binucleine 2 (EMD Millipore) for 10 min before being processed for immunostaining. For pT182-Polo signal detection, cells were treated with 100 nM okadaic acid (BioShop Canada Inc.) for 1 h before lysis.
Immunofluorescence and Western blotting
Primary antibodies used in immunofluorescence and Western blotting were anti–α-tubulin DM1A from mouse (Sigma-Aldrich), anti-Myc 9E10 from mouse (Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-Polo MA294 (a gift of D. Glover, University of Cambridge, Cambridge, England, UK), anti-pT210 Plk1 2A3 from mouse (recognizes Drosophila pT182 Polo; Abcam), rabbit anti-Map205 (a gift from A. Pereira, University of Massachusetts Medical School, Worcester, MA), anti-GFP from rabbit (#A6455; Invitrogen), anti-GST from rabbit (#2622; Cell Signaling Technology), and peroxidase-conjugated ChromPure rabbit IgG (for PrA detection; Jackson ImmunoResearch Laboratories, Inc.).
For immunofluorescence, cells were fixed with 4% formaldehyde for 10 min, then permeabilized and blocked in PBS containing 0.1% Triton X-100 and 1% BSA (PBSTB). Cells were incubated with primary antibodies diluted in PBSTB for 2 h at RT, washed three times in PBS, and incubated with secondary antibodies diluted in PBSTB for 1 h at RT. Cells were washed three times in PBS before being mounted in Vectashield medium with DAPI (Vector Laboratories).
Protein affinity purification and immunoprecipitation
Protein A affinity purifications were performed essentially as described previously (D’Avino et al., 2009). In brief, around 10 million cells were harvested and resuspended in 400 µl of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 1 mM EDTA, and 10% glycerol) supplemented with protease inhibitors (1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin). Lysates were clarified by centrifugation at 14,000 rpm for 10 min in a tabletop centrifuge at 4°C. Supernatants were incubated for 1 h at 4°C with 20 µl of DynaBeads (Invitrogen) that we previously conjugated to rabbit IgG. Beads were washed five times with 1 ml of lysis buffer for 5 min at 4°C. Purification products were eluted by heating at 95°C for 5 min in 20 µl of Laemmli buffer twice (Sigma-Aldrich) and analyzed by Western blotting.
For immunoprecipitation of Polo-Myc (WT, T182A, and T182D), extracts were prepared as for Protein A affinity purifications and lysates were incubated with anti-Myc antibodies for 1 h at 4°C, then incubated with 20 µl of Protein G–conjugated Dynabeads (Life Technologies) for 45 min at 4°C, before being washed in lysis buffer.
Transient transfections
Transfections of D-Mel2 cells with plasmids were performed using X-tremeGENE HP DNA Transfection Reagent (Roche) according to the manufacturer’s instructions. For RNA interference, cells were transfected in 6-well plates with 20 µg of dsRNA (Polo 3′ UTR dsRNA or Map205 dsRNA) using Transfast reagent (Promega). The control dsRNA was generated against the sequence of the bacterial kanamycin resistance gene. Cells were analyzed between 24 h and 72 h later by immunoblotting, immunofluorescence, or live cell imaging.
GST pull-down assay
For pull-down assays, pelleted cells transfected with Polo-Myc (WT, T182A, and T182D) from confluent 75-cm2 flasks were resuspended in lysis buffer (75 mM K-Hepes, pH 7.5, 150 mM KCl, 2 mM EGTA, 2 mM MgCl2, 5% glycerol, 0.2% Triton X-100, 1 mM DTT, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin) and centrifuged for 15 min at 4°C. Clarified lysates were incubated with GST-Map205254-416, GST-Map205254-375, or GST Sepharose beads for 2 h at 4°C. Beads were washed five times with lysis buffer before SDS-PAGE and immunoblotting.
Polo kinase expression and purification
The coding region of Drosophila Polo was amplified by PCR and cloned into pFastbac-Htb vector. To express Polo protein, a bacmid was produced and transfected into Sf9 cells to generate the baculovirus according to the manufacturer’s protocol (Bac-to-Bac; Invitrogen). The virus was subsequently amplified to infect a larger quantity of cells (400 ml) in a spinner flask. 72 h after infection, cells were collected, washed with 1× PBS, and frozen until purification. To purify the recombinant protein, the Sf9 cell pellet was first lysed in lysis buffer (50 mM Tris, pH 7.5, 300 mM KCl, 10 mM imidazole, 0.5% Triton X-100, 10 mM β-Mercaptoethanol, 1 mM PMSF, 10 µg/ml of leupeptin, pepstatin A, and chymostatin). The lysate was then spun at 45,000 rpm in a rotor (T-865; Sorvall) at 4°C for 40 min. The supernatant was recovered and allowed to bind to Talon resins (Takara Bio Inc.) preequilibrated with lysis buffer for 1 h at 4°C. Affinity resin-bound proteins were washed thoroughly with lysis buffer with 600 mM KCl and eluted with imidazole-containing buffer (50 mM Tris, pH 7.5, 250 mM imidazole, 300 mM KCl, and 1 mM DTT). Purified Polo was then dialyzed into storage buffer (50 mM Tris, pH 7.5, 300 mM KCl, 1 mM DTT, and 20% glycerol) overnight before flash freezing in small aliquots in liquid nitrogen.
In vitro binding assay and kinase assays
Purified GST-Map205254-416 were incubated with active purified Polo or hPlk1 or immunoprecipitated Polo-Myc (WT, T182A, T182D) in binding buffer (75 mM K-Hepes, pH 7.5, 150 mM KCl, 2 mM EGTA, 2 mM MgCl2, 5% glycerol, 0.2% Triton X-100, 1 mM DTT, 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin) for 1 h at 4°C.
For kinase assays, reactions were performed in kinase buffer (20 mM K-Hepes, pH 7.5, 2 mM MgCl2, and 1 mM DTT) with 0.5 µM ATP, [32P]γ-ATP, and 1 µg casein at 30°C for 15 min. For Polo inhibition in the kinase assay, BI 2536 was added at 300 nM. Reactions were stopped with the addition of the Laemmli buffer and heating at 95°C for 2 min. Samples were separated by SDS-PAGE and transferred onto nitrocellulose for autoradiography and Western blotting.
Microscopy
Images of fixed cells were acquired on an microscope (AxioImager) with a 100× oil objective lens (NA 1.4) and an AxioCam HRm camera, using AxioVision software (all from Carl Zeiss). Line-scan measurements of GFP intensity along the intercellular bridge were generated using ImageJ software. For time-lapse microscopy, D-Mel2 Cells stably expressing PoloWT-GFP, PoloT182A-GFP, and PoloT182D-GFP were cultured in optical glass-bottom plate (Greiner Bio-One). Cells were treated for 24 h with dsRNA targeting the 3′ UTR of endogenous Polo or control dsRNA. Live imaging shown in Fig. 1 (and Videos 1 and 2) was performed using a spinning disc confocal system (Ultraview Vox; PerkinElmer), a Plan-Apochromat 100× oil immersion objective lens (NA 1.4), and an Orca-R2 charge-coupled device (CCD) camera (Hamamatsu Photonics) with 2 × 2 binning, using Volocity 6.0 software (PerkinElmer). Binucleine 2 was added at anaphase onset. Live imaging shown in Fig. 4 was performed using a DeltaVision elite microscope (Applied Precision) and a 60× oil objective lens (NA 1.42) in a temperature-controlled environment (25°C). Images were acquired using a CoolSnap HQ2 camera (Photometrics). For each condition in this experiment, a minimum of three independent experiments was performed, a minimum of 91 cell divisions was recorded, and cytokinesis defects were counted manually. Images were treated using SoftWoRx (Applied Precision), ImageJ, and Photoshop (Adobe) software.
Online supplemental material
Fig. S1 shows additional experiments and controls accompanying Fig. 1. Fig. S2 shows that polo hyperphosphorylation at Thr182 induced by Aurora B overexpression depends on Aurora B kinase activity. Fig. S3 shows that Map205 inhibits the activity of Drosophila Polo and human Plk1. Video 1 shows time-lapse imaging of a Polo-GFP–expressing cell in mitosis. Video 2 shows time-lapse imaging of a Binucleine 2–treated Polo-GFP–expressing cell in mitosis. Video 3 shows time-lapse imaging of a cell depleted of endogenous Polo and expressing PoloWT-GFP. Video 4 shows time-lapse imaging of a cell depleted of endogenous Polo and expressing PoloT182A-GFP showing an early cytokinesis failure. Video 5 shows time-lapse imaging of a cell depleted of endogenous Polo and expressing PoloT182A-GFP showing a late cytokinesis failure. Video 6 shows time-lapse imaging of a cell depleted of endogenous Polo and expressing PoloT182D-GFP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408081/DC1.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to V. Archambault, G.R. Hickson, and B.H. Kwok, and from the Natural Sciences and Engineering Research Council of Canada to S. Carréno. V. Archambault, B.H. Kwok, and S. Carréno hold New Investigator Awards from the CIHR. V. Archambault, B.H. Kwok, S. Carréno, and G.R. Hickson were supported by Junior 1 salary awards from the Fonds de recherche du Québec – Santé (FRQS). D. Kachaner is a 2013 recipient of a Prix pour les Jeunes Chercheurs from the Bettencourt Schueller Foundation and holds a postdoctoral fellowship from the Cole Foundation. K.B. El Kadhi and G. Lépine hold a studentship from the FRQS. Institut de Recherche en Immunologie et en Cancérologie (IRIC) is supported in part by the Canadian Center of Excellence in Commercialization and Research (CECR), the Canada Foundation for Innovation (CFI), and by the FRQS. G.R. Hickson is also supported by the CFI.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- KD
- kinase domain
- PBD
- Polo-Box domain
- Plk
- Polo-like kinase
- WT
- wild type
References
- Archambault, V., and Carmena M.. 2012. Polo-like kinase-activating kinases: Aurora A, Aurora B and what else? Cell Cycle. 11:1490–1495. 10.4161/cc.19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault, V., and Glover D.M.. 2009. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10:265–275. 10.1038/nrm2653 [DOI] [PubMed] [Google Scholar]
- Archambault, V., D’Avino P.P., Deery M.J., Lilley K.S., and Glover D.M.. 2008. Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev. 22:2707–2720. 10.1101/gad.486808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, I.M., Peters U., Kapoor T.M., and Straight A.F.. 2007. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS ONE. 2:e409. 10.1371/journal.pone.0000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma, W., Raaijmakers J.A., and Medema R.H.. 2012. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 37:534–542. 10.1016/j.tibs.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Bruinsma, W., Macurek L., Freire R., Lindqvist A., and Medema R.H.. 2014. Bora and Aurora-A continue to activate Plk1 in mitosis. J. Cell Sci. 127:801–811. 10.1242/jcs.137216 [DOI] [PubMed] [Google Scholar]
- Burkard, M.E., Randall C.L., Larochelle S., Zhang C., Shokat K.M., Fisher R.P., and Jallepalli P.V.. 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 104:4383–4388. 10.1073/pnas.0701140104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, M., Pinson X., Platani M., Salloum Z., Xu Z., Clark A., Macisaac F., Ogawa H., Eggert U., Glover D.M., et al. 2012a. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 10:e1001250. 10.1371/journal.pbio.1001250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012b. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E.H., Santamaria A., Silljé H.H., and Nigg E.A.. 2008. Plk1 regulates mitotic Aurora A function through βTrCP-dependent degradation of hBora. Chromosoma. 117:457–469. 10.1007/s00412-008-0165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino, P.P., Archambault V., Przewloka M.R., Zhang W., Lilley K.S., Laue E., and Glover D.M.. 2007. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS ONE. 2:e572. 10.1371/journal.pone.0000572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino, P.P., Archambault V., Przewloka M.R., Zhang W., Laue E.D., and Glover D.M.. 2009. Isolation of protein complexes involved in mitosis and cytokinesis from Drosophila cultured cells. Methods Mol. Biol. 545:99–112. 10.1007/978-1-60327-993-2_6 [DOI] [PubMed] [Google Scholar]
- Elia, A.E., Cantley L.C., and Yaffe M.B.. 2003a. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 299:1228–1231. 10.1126/science.1079079 [DOI] [PubMed] [Google Scholar]
- Elia, A.E., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., and Yaffe M.B.. 2003b. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 115:83–95. 10.1016/S0092-8674(03)00725-6 [DOI] [PubMed] [Google Scholar]
- Fenton, B., and Glover D.M.. 1993. A conserved mitotic kinase active at late anaphase-telophase in syncytial Drosophila embryos. Nature. 363:637–640. 10.1038/363637a0 [DOI] [PubMed] [Google Scholar]
- Fuller, B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., and Kapoor T.M.. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136. 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, Y.J., Lin C.Y., Ma S., and Erikson R.L.. 2002a. Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc. Natl. Acad. Sci. USA. 99:1984–1989. 10.1073/pnas.042689299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, Y.J., Ma S., Terada Y., and Erikson R.L.. 2002b. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 277:44115–44120. 10.1074/jbc.M202172200 [DOI] [PubMed] [Google Scholar]
- Kachaner, D., Filipe J., Laplantine E., Bauch A., Bennett K.L., Superti-Furga G., Israël A., and Weil R.. 2012. Plk1-dependent phosphorylation of optineurin provides a negative feedback mechanism for mitotic progression. Mol. Cell. 45:553–566. 10.1016/j.molcel.2011.12.030 [DOI] [PubMed] [Google Scholar]
- Kothe, M., Kohls D., Low S., Coli R., Rennie G.R., Feru F., Kuhn C., and Ding Y.H.. 2007. Selectivity-determining residues in Plk1. Chem. Biol. Drug Des. 70:540–546. 10.1111/j.1747-0285.2007.00594.x [DOI] [PubMed] [Google Scholar]
- Lowery, D.M., Lim D., and Yaffe M.B.. 2005. Structure and function of Polo-like kinases. Oncogene. 24:248–259. 10.1038/sj.onc.1208280 [DOI] [PubMed] [Google Scholar]
- Macůrek, L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., and Medema R.H.. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 455:119–123. 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Neef, R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., and Barr F.A.. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444. 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Park, J.E., Soung N.K., Johmura Y., Kang Y.H., Liao C., Lee K.H., Park C.H., Nicklaus M.C., and Lee K.S.. 2010. Polo-box domain: a versatile mediator of polo-like kinase function. Cell. Mol. Life Sci. 67:1957–1970. 10.1007/s00018-010-0279-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, A., Doshen J., Tanaka E., and Goldstein L.S.. 1992. Genetic analysis of a Drosophila microtubule-associated protein. J. Cell Biol. 116:377–383. 10.1083/jcb.116.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki, M., Glotzer M., Kraut N., and Peters J.M.. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 12:713–725. 10.1016/j.devcel.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Petronczki, M., Lénárt P., and Peters J.M.. 2008. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell. 14:646–659. 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Qian, Y.W., Erikson E., and Maller J.L.. 1999. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol. Cell. Biol. 19:8625–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud, S., Carmena M., and Earnshaw W.C.. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812. 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Santamaria, A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., et al. 2007. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell. 18:4024–4036. 10.1091/mbc.E07-05-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, A., Coppinger J.A., Du H., Jang C.Y., Yates J.R. III, and Fang G.. 2008a. Plk1- and β-TrCP-dependent degradation of Bora controls mitotic progression. J. Cell Biol. 181:65–78. 10.1083/jcb.200712027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, A., Coppinger J.A., Jang C.Y., Yates J.R., and Fang G.. 2008b. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658. 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurnyy, Y., Toms A.V., Hickson G.R., Eck M.J., and Eggert U.S.. 2010. Binucleine 2, an isoform-specific inhibitor of Drosophila Aurora B kinase, provides insights into the mechanism of cytokinesis. ACS Chem. Biol. 5:1015–1020. 10.1021/cb1001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier, M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., et al. 2007. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17:316–322. 10.1016/j.cub.2006.12.037 [DOI] [PubMed] [Google Scholar]
- Wolfe, B.A., Takaki T., Petronczki M., and Glotzer M.. 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7:e1000110. 10.1371/journal.pbio.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y., Takeo S., Florens L., Hughes S.E., Huo L.J., Gilliland W.D., Swanson S.K., Teeter K., Schwartz J.W., Washburn M.P., et al. 2007. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 5:e323. 10.1371/journal.pbio.0050323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Shen C., Wang T., and Quan J.. 2013. Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20:1047–1053. 10.1038/nsmb.2623 [DOI] [PubMed] [Google Scholar]
- Zitouni, S., Nabais C., Jana S.C., Guerrero A., and Bettencourt-Dias M.. 2014. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15:433–452. 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.