Abstract
Background
WNK1 (With No-lysine Kinase 1) modulates numerous sodium transport-related ion channels involved in regulation of blood pressure. Several studies have indicated associations between the common variants of the WNK1 gene and hypertension or blood pressure levels. However, little data exists on Asian populations and normotensive or pre-hypertensive subjects. Our aim was to detect whether the common variations in the WNK1 gene are potential contributors to individual variations in blood pressure in a family-based sample.
Material/Methods
525 individuals from 116 families were selected from a rural community of Northern China. Five single-nucleotide polymorphisms were selected from the WNK1 gene. Single-marker and haplotype analyses were conducted using the Family-Based Association Test program.
Results
Regretful, no associations for the 5 WNK1 SNPs and the constructed haplotype blocks of WNK1 with blood pressure level reached nominal statistical significance.
Conclusions
We conclude that although multiple candidate genes are involved in development of hypertension, the genetic polymorphism in WNK1 is not a major contributor to the observed variability in blood pressure and familial clustering risk of hypertension.
MeSH Keywords: Blood Pressure, Family, Polymorphism, Genetic
Background
Hypertension is a global public health challenge because of its high prevalence and the concomitant increase in risk of vascular disease [1–3]. An abundance of evidence has confirmed that an individual’s genetic profile may play a large role in blood pressure regulation, and hypertension is recognized as a polygenic syndrome [4,5]. Improved understanding of mechanisms of BP regulation should facilitate advances in the prevention and treatment of hypertension. Because BP is a heritable trait, efforts are underway to identify genetic variants that have a role in BP regulation [6–11].
WNK1 (With No-lysine Kinase 1) is a serine-threonine kinase regulating numerous ion channels involved in sodium and potassium transport [12–17]. WNK1 maps to chromosome 12p13.3, is encoded by 28 exons, and spans over 150 kb of genomic DNA. It is ubiquitously expressed in various tissues, particularly in the kidney and cardiovascular system. WNK1 have been implicated as important modulators of salt homeostasis, regulating the balance between renal sodium reabsorption and potassium excretion. Mutations in WNK1 cause pseudohypoaldosteronism type 2 (PHA2), a rare autosomal-dominant disorder primarily characterized by early-onset hypertension and hyperkalemia [18]. Therefore, the possibility has been proposed that genetic mutations in WNK1 affect blood pressure variations and/or susceptibility to essential hypertension.
Indeed, accumulated evidence has indicated associations between the common variants of the WNK1 gene and blood pressure levels [19–23]. However, these previous studies always emphasized the relationship in hypertensive subjects but not in normotensive or pre-hypertensive subjects, and the data in Asian populations are rare. In the present study, we investigated whether the common variations in the WNK1 gene are potential contributors to individual variations in blood pressure in a family-based sample of 525 normotensive or pre-hypertensive subjects from northern China.
Material and Methods
Subjects
In northern China, a community-based BP screening was conducted among persons aged 18–60 years in the study villages to identify potential probands and their families for our study. All the study subjects belong to the Chinese Han ethnic group. We recruited subjects with mean systolic BP (SBP) between 130 and 160 mm Hg and/or diastolic BP (DBP) between 85 and 100 mm Hg and no use of antihypertensive medications, and their siblings, spouses, and children. All subjects underwent clinical and biochemical investigation to exclude those with secondary hypertension, chronic kidney disease, or type 2 diabetes mellitus. The institutional ethics committee of Xi’an Jiaotong University Medical School approved the study protocol, and written informed consent for the total program was obtained from each participant. All of the procedures were performed in accordance with institutional guidelines.
BP Measurement
Three random-zero BP measurements were obtained using a Hawksley random-zero sphygmomanometer (Hawksley & Sons Ltd, Lancing, UK; zero range 0–20 mmHg) with a 1-minute interval. BP was measured by trained and certified observers according to a common protocol adapted from procedures recommended by the American Heart Association. BP was measured with the participant in sitting position after 5 min of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 min prior to their BP measurement. Systolic BP (SBP) and diastolic BP (DBP) were determined as the first and fifth phases of the Korotkoff sounds, respectively. MBP was defined as:
DNA extraction and genotyping
Peripheral venous blood was drawn from each participant. Genomic DNA was extracted from whole blood using the Maxwell 16 DNA Purification Kit (Promega Corporation, Madison, WI). The choice of SNP was informed by the results of a prior analysis of WNK1. All the genotyping experiments were done by the Shanghai Generay Biotech Co., Ltd. (http://www.generay.com.cn/) using ligase detection reactions (LDR). The target DNA sequences were amplified using a multiplex PCR method. After the completion of the amplification, 1 μl of proteinase K (20 mg/ml) was added, then heated at 70°C for 10 min and quenched at 94°C for 15 min. The ligation reaction for each subject was carried out in a final volume of 10 μl containing 2 μl of multi-PCR product, 1 μl of probe, 0.125 μl of 40 U/μl Taq DNA ligase (NEB, USA), 1 μl of 10× Taq DNA ligase buffer, and 6 μl H2O. The LDR was performed using 25 cycles of 94°C for 30 sec and 55°C for 4 min. The fluorescent products of LDR were differentiated by use of an ABI sequencer 377. Additionally, about 5% of the samples were randomly selected and retested by direct DNA sequencing on a 3730xl DNA analyzer (Applied Biosystems) and the results were 100% concordant.
Statistical analysis
The Mendelian consistency of the SNP genotype data was assessed by PLINK and PedCheck on parental SNP data (Figure 1). Departure from Hardy-Weinberg equilibrium was tested with χ2 test on parental SNP data. We used Haploview software (version 4.0, http://www.broad.mit.edu/mpg/haploview) to estimate the extent of pairwise linkage disequilibrium between SNPs. We used the Family-Based Association Test (FBAT) program (version 2.0.2, http://www.biostat.harvard.edu/fbat/default.html) to test the association of single-marker and haplotypes with adjusted phenotypes [24]. Three genetic models (additive, dominant, and recessive) were tested. To assess the effect of genetic variants on the trait value, a univariate FBAT test was performed for each allele and haplotype. This test provides a z-statistic with its corresponding P value. The false discovery rate (FDR) method was used to correct for multiple testing.
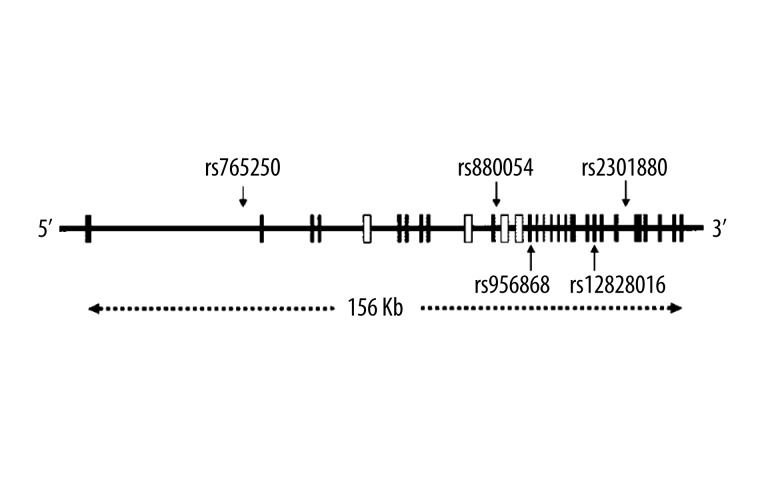
Figure 1.
Structure of the human WNK1 gene and location of the SNPs analyzed. Exons are shown as vertical lines. The locations of the 5 genotyped SNPs are indicated by arrows.
Results
Characteristics of the study participants
We recruited 525 individuals from 116 families, including 23 families with 3 generations of pedigree and 93 families with 2 generations of pedigree. All the families had 2 or more children. Of these 525 individuals, 481 had normal blood pressure and 44 were essential hypertensive patients. The characteristics of the study individuals are given in Table 1. The probands had higher mean baseline levels of SBP and DBP than their siblings, spouses, and children, whereas the parents had the highest SBP levels among all of the groups.
Table 1.
Characteristics of the study participants at the baseline examination.
| Probands | Siblings | Spouses | Offspring | Parents | |
|---|---|---|---|---|---|
| No. of participants | 102 | 172 | 18 | 47 | 186 |
| Age (years) | 40.5±7.6 | 39.4±7.3 | 47.4±6.1 | 23.3±7.1 | 66.1±8.2 |
| Male (%) | 69.6 | 50 | 22.2 | 46.8 | 47.8 |
| Body mass index, kg/m2 | 23.0±2.8 | 22.3±2.9 | 22.5±3.2 | 19.9±2.5 | 20.4±2.6 |
| Systolic, mmHg | 120.6±11.0 | 107.4±10.5 | 108.6±12.2 | 102.4±10.7 | 123.6±21.6 |
| Diastolic, mmHg | 78.8±7.9 | 70.0±8.2 | 70.6±6.9 | 62.9±8.7 | 70.7±10.8 |
| Mean arterial pressure, mmHg | 92.7±8.2 | 82.4±8.5 | 83.3±7.9 | 76.1±9.0 | 88.3±13.4 |
| 24 h urinary sodium, mmol | 228±11.6 | 213±16.4 | 218±20.8 | 115±23.4 | 124±21.4 |
| 24 h urinary potassium, mmol | 36.8±6.83 | 41.1±9.13 | 39.4±12.3 | 45.6±18.7 | 47.3±17.1 |
Continuous variables are indicated as mean ±SD. BP, blood pressure.
Allele frequencies and Hardy-Weinberg equilibrium test
Table 2 summarizes the genomic location, allele frequency, and Hardy-Weinberg test results for the 5 WNK1 SNPs analyzed. None of the SNPs showed statistically significant deviation from Hardy-Weinberg equilibrium.
Table 2.
Description of the WNK1 SNPs genotyped.
| WNK1 SNP | WNK1 position | Alleles | Minor-Allele frequency* | Hardy-Weinberg Test* | |
|---|---|---|---|---|---|
| Pearson’s χ2 | P value | ||||
| rs880054 | Intron 10 | A/G | 0.27 | 0.72 | 0.396 |
| rs12828016 | Exon 21 | Met/lle | 0.25 | 0.022 | 0.883 |
| rs956868 | Exon 13 | Pro/Thr | 0.21 | 1.833 | 0.176 |
| rs2301880 | Intron 23 | C/T | 0.21 | 0.029 | 0.865 |
| rs765250 | Intron 1 | A/G | 0.12 | 0.004 | 0.949 |
Minor allele frequency and Hardy-Weinberg tests relate to the parents (parental generation).
Association tests between WNK1 and blood pressure level
As shown in Table 3, single-locus analyses conducted with the use of FBAT revealed no significant evidence of an association of blood pressure level with the 5 common variation in WNK1 (all P>0.05). To further increase the statistical power of the study, we also applied the 3 genetic models and false discovery rate (FDR) method, but we still found no significant differences.
Table 3.
Association between WNK1 SNPs and blood pressure.
| SNP | Allele | SBP | DBP | MAP | |||
|---|---|---|---|---|---|---|---|
| z | P | z | P | z | P | ||
| rs880054 | A | 0.154 | 0.878 | 0.458 | 0.647 | 0.340 | 0.733 |
| rs12828016 | G | 0.041 | 0.967 | 0.420 | 0.674 | 0.274 | 0.784 |
| rs956868 | A | 0.861 | 0.389 | 0.941 | 0.346 | 0.951 | 0.342 |
| rs2301880 | C | 0.174 | 0.860 | 0.696 | 0.636 | 0.302 | 0.762 |
| rs765250 | A | 0.897 | 0.369 | 0.913 | 0.361 | 1.014 | 0.311 |
z indicates test statistic for FBAT. BP – blood pressure; FBAT – Family Based Association Test; MAP – mean arterial pressure; SNP – single nucleotide polymorphism, P values are corrected for multiple testing (FDR <0.05). FDR – false discovery rate.
Because all the SNPs within the same gene had been found to be in strong LD, we also constructed haplotype blocks of WNK1 SNPs by using Haploview software, and found that the 5 SNPs were located in 1 haploblock. However, no significant differences in haplotype distributions were found to be associated with blood pressure (P>0.05).
Discussion
We investigated the association of 5 common SNPs with blood pressure in a family-based sample of 528 normotensive or pre-hypertensive subjects from northern China. However, in the current study, despite using data corrected for multiple testing, we still failed to detect any significant associations. Furthermore, we also found no significant association of their haplotypes with blood pressure.
Advances in human genetics have provided a number of opportunities for investigating the association between genetics and disease, but also a growing recognition that results of many published associations failed to be replicated. A number of studies have found that WNK1 plays a vital role in maintaining sodium and potassium homeostasis in the kidney, and consequently blood pressure regulation. On the basis of in vitro and animal studies, WNK1 seemed to be an obvious candidate gene to test for associations with blood pressure [13–17]. Newhouse et al. tested for associations among 19 WNK1 SNPs and EH in 712 severely hypertensive families, and observed evidence suggesting an association between variants of WNK1 and severity of hypertension [22]. Moreover, Tobin et al. also reported that rs880054 in WNK1 contributes to BP variation in a population-based sample of 996 subjects from 250 white European families [21]. Recently, Osada et al. reported that the tSNPs rs880054, rs956868, and rs12828016 in the WNK1 gene were associated with blood pressure variations in the general Japanese population, and that the constructed haplotypes were associated with Na/K intake ratio [25]. However, through careful observation of these positive outcomes between tSNP and blood pressure, we found that the association was inconsistent or nominal, such as with rins880054, rs956868, and rs765250, in which findings of the same relationship were not replicated in other studies. Furthermore, Kokubo et al. tested for association among 7 WNK1 SNPs and EH in 1818 Japanese individuals, 771 of whom were hypertensive, but no association was found [26]. Several researchers have criticized the above-mentioned studies for not conducting any haplotype analysis or considering quantitative BP phenotypes in association tests with WNK1. Of course, different ethnicity may be an irreconcilable factor because these populations show disparate minor allele frequencies in the 5 tSNPs. The current study, in spite of using a family-based sample and quantitative BP phenotypes, still failed to find any association of tSNP or haplotypes with blood pressure, which is in accord with Kokubo’s conclusion. Therefore, we believe the variation at WNK1 does not play a major role in the risk of hypertension.
Our study has several important strengths and limitations. The subjects were recruited from several neighboring rural communities that were similar with respect to lifestyle and environmental risk factors, including diet and physical activity. Furthermore, because this study was based on the family pedigree, the within-family association test can eliminate the effect of an admixed and stratified population using FBAT. Thus, confounding of genetic associations due to these factors should have been minimal. In contrast, because all of the subjects were recruited from the northern Chinese population, our results will require replication in other cohorts to determine generalizability to other ethnicities and to populations with different dietary habits. Moreover, the sample size in the current study was small, so we need to repeat the study using a larger sample.
Conclusions
We did not find a significant association between each of the 5 tagging SNPs and haplotypes in WNK1 and blood pressure in a northern Chinese population. These results strongly support the view that variation at WNK1 does not play a major role in the familial clustering risk of hypertension. However, we cannot exclude that regulatory variation that is distinct from WNK1 and not in linkage disequilibrium with any of the variants tested here might affect the expression of this gene and influence blood pressure.
Footnotes
Source of support: This study was supported by grant 2012CB517804 from the National Program on Key Basic Research Project of China (973 Program) and grants 81200512 (to Liu), 81070218 and 8130357 (to Mu) from the Natural Science Foundation of China
Competing interests
The authors declare that they have no competing interests.
References
- 1.Collaboration PS. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360(9349):1903–13. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 2.Lawes C, Hoorn SV, Rodgers A. Global burden of blood-pressure-related disease. Lancet. 2008;371(9623):1513–18. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 3.Staessen JA, Wang J, Bianchi G, Birkenhäger WH. Essential hypertension. Lancet. 2003;361(9369):1629–41. doi: 10.1016/S0140-6736(03)13302-8. [DOI] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular Mechanisms of Human Hypertension. Cell. 2001;104(4):545–56. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Oparil S, Zaman MA, Calhoun DA. Pathogenesis of hypertension. Ann Intern Med. 2003;139:761–76. doi: 10.7326/0003-4819-139-9-200311040-00011. [DOI] [PubMed] [Google Scholar]
- 6.Luft FC. Present status of genetic mechanisms in hypertension. Med Clin North Am. 2004;88(1):1–18. doi: 10.1016/s0025-7125(03)00118-4. [DOI] [PubMed] [Google Scholar]
- 7.Caulfield M, Newhouse S, Munroe P. Genetics of essential hypertension Principles and Practice of Clinical Cardiovascular Genetics. Oxford University Press; Oxford, UK: 2010. pp. 329–36. [Google Scholar]
- 8.Franceschini N, Reiner AP, Heiss G. Recent findings in the genetics of blood pressure and hypertension traits. Am J Hypertens. 2011;24(4):392–400. doi: 10.1038/ajh.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tobin MD, Tomaszewski M, Braund PS, et al. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51(6):1658–64. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 10.Armani C, Botto N, G Andreassi M. Susceptibility genes in hypertension. Curr Pharm Des. 2011;17(28):2973–86. doi: 10.2174/138161211798157667. [DOI] [PubMed] [Google Scholar]
- 11.Määttä K, Kunnas T, Nikkari ST. Contribution of SLC7A1 genetic variant to hypertension, the TAMRISK study. BMC Med Gen. 2013;14(1):69–72. doi: 10.1186/1471-2350-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naray-Fejes-Toth A, Snyder PM, Fejes-Toth G. The kidney-specific WNK1 isoform is induced by aldosterone and stimulates epithelial sodium channel-mediated Na+ transport. Proc Natl Acad Sci USA. 2004;101:17434–39. doi: 10.1073/pnas.0408146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahle KT, Ring AM, Lifton RP. Molecular Physiology of the WNK Kinases. Annu Rev Physiol. 2008;70:329–55. doi: 10.1146/annurev.physiol.70.113006.100651. [DOI] [PubMed] [Google Scholar]
- 14.Huang CL, Kuo E. Mechanisms of Disease: WNK-ing at the mechanism of salt-sensitive hypertension. Nature Clinical Practice Nephrology. 2007;11:623–30. doi: 10.1038/ncpneph0638. [DOI] [PubMed] [Google Scholar]
- 15.Hoorn EJ, Lubbe N, Zietse R. The renal WNK kinase pathway: a new link to hypertension. Nephrol Dial Transplant. 2009;24:1074–77. doi: 10.1093/ndt/gfp013. [DOI] [PubMed] [Google Scholar]
- 16.O’Reilly M, Marshall E, Speirs HJ, Brown RW. WNK1, a gene within a novel blood pressure control pathway, tissue-specifically generates radically different isoforms with and without a kinase domain. J Am Soc Nephrol. 2003;14:2447–56. doi: 10.1097/01.asn.0000089830.97681.3b. [DOI] [PubMed] [Google Scholar]
- 17.Fang L, Liu J, Li D, et al. WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci USA. 2006;103(22):8558–63. doi: 10.1073/pnas.0603109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson FH, Disse-Nicodeme S, Choate KA, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–12. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 19.Cun Y, Li J, Tang W, et al. Association of WNK1 exon 1 polymorphisms with essential hypertension in Hani and Yi minorities of China. J Genet Genomics. 2011;38(4):165–71. doi: 10.1016/j.jgg.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Putku M, Kepp K, Sober S, et al. Novel polymorphic AluYb8 insertion in the WNK1 gene is associated with blood pressure variation in Europeans. Hum Mutat. 2011;32(7):806–14. doi: 10.1002/humu.21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobin MD, Timpson NJ, Wain LV, et al. Common Variation in the WNK1 Gene and Blood Pressure in Childhood: The Avon Longitudinal Study of Parents and Children. Hypertension. 2008;52:974–79. doi: 10.1161/HYPERTENSIONAHA.108.118414. [DOI] [PubMed] [Google Scholar]
- 22.Tobin MD, Raleigh SM, Newhouse S, et al. Association of WNK1 Gene Polymorphisms and Haplotypes With Ambulatory Blood Pressure in the General Population. Circulation. 2005;112:3423–29. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 23.Newhouse SJ, Wallace C, Dobson R, et al. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Human Molecular Genetics. 2005;14(13):1805–14. doi: 10.1093/hmg/ddi187. [DOI] [PubMed] [Google Scholar]
- 24.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype – phenotype associations. Eur J Hum Genet. 2001;9(4):301–6. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 25.Osada Y, Miyauchi R, Goda T, et al. Variations in the WNK1 gene modulates the effect of dietary intake of sodium and potassium on blood pressure determination. J Human Genet. 2009;54:474–78. doi: 10.1038/jhg.2009.64. [DOI] [PubMed] [Google Scholar]
- 26.Kokubo Y, Kamide K, Inamoto N, et al. Identification of 108 SNPs in TSC, WNK1, and WNK4 and their association with hypertension in a Japanese general population. J Hum Genet. 2004;49:507–15. doi: 10.1007/s10038-004-0181-0. [DOI] [PubMed] [Google Scholar]