Figure 3.
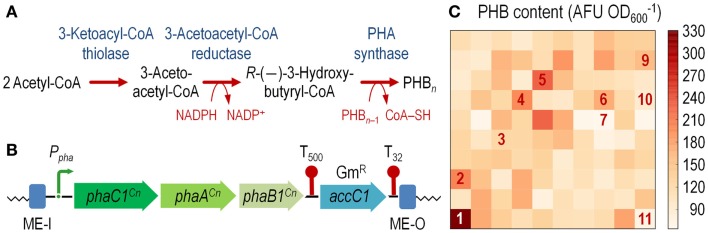
Construction of an E. coli cell factory expressing the phaC1AB1 gene cluster from C. necator as a chromosomal insertion. (A) Poly(3-hydroxybutyrate) (PHB) biosynthesis pathway. Three enzymes are necessary for de novo synthesis of PHB in C. necator: a 3-ketoacyl-coenzyme A (CoA) thiolase (PhaA), a NADPH-dependent 3-acetoacetyl-CoA reductase (PhaB1), and a PHB synthase (PhaC1). PhaA and PhaB1 catalyze the condensation of two molecules of acetyl-CoA to 3-acetoacetyl-CoA and the reduction of acetoacetyl-CoA to R-(–)-3-hydroxybutyryl-CoA, respectively. PhaC1 polymerizes these monomers to PHB, whereas one CoA-SH molecule per monomer is released. The resulting PHB polymer is stored as water-insoluble granules in the cytoplasm of the cells. (B) Organization of the functional elements borne by plasmid pBAM1-6-pha and transferred into the chromosome of the recipient E. coli strain. The transcriptional terminators included in the plasmid backbone, which flank the gentamicin resistance (GmR) determinant (accC1), are depicted as T500 and T32. Note that the elements in this outline are not drawn to scale. (C) Exploring the landscape of PHB synthesis in E. coli transconjugants. The phaC1AB1 gene cluster from C. necator was randomly integrated into the chromosome of E. coli JW2293-1 (Δpta), and 24-h cultures of individual colonies were analyzed for PHB accumulation by fluorimetry after staining the cells with Nile red (see Materials and Methods for details). Several colonies, identified by numbers in the heat-map, were kept and further analyzed to establish the precise site of mini-Tn5(phaC1AB1) insertion (Table S2 in the Supplementary Material). AFU, arbitrary fluorescence units.