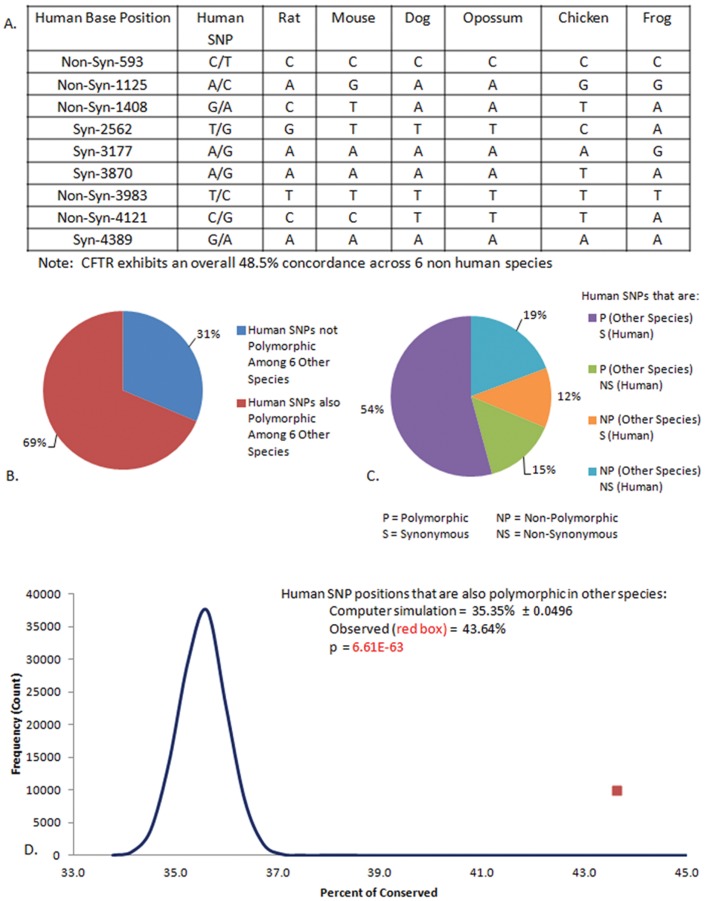
Figure 4. Positions exhibiting polymorphism in human CFTR are also polymorphic among other species.
A: Six of nine CFTR coding SNPs identified by unbiased analysis of individuals in 1000 Genomes were also were polymorphic among diverse species, despite approximately 50% overall nucleotide identity among the non-human CFTRs being analyzed. B, C: CFTR and 21 other genes (Table S3) were investigated in the same fashion shown in Panel A. The majority of SNPs in exonic regions found to be polymorphic were synonymous (p = 2.7×10−9, versus the stochastic ratio otherwise expected for non-synonymous to synonymous polymorphism). In order to increase stringency, only those genes in Figure 2A with ≥50% concordance across the six non-human species were included in the analysis. D. CFTR homologs in four evolutionarily distant species (horse, frog, zebrafish, and shark) were aligned with the human coding strand, both independently and collectively. In the collective alignment, ∼43% of the coding sequence was invariant. A computer simulation was conducted and the total number of differences from human placed randomly within the human CFTR reading frame of 4443 bp. The goal was to determine in a conservative fashion whether concordance observed in a multiple species alignment could be accounted for by chance. The simulation was performed 120,000 times and the numbers of differences from human tabulated. The mean concordance (35.4%) and standard deviation (∼0.05) for this set of simulation data was calculated and differed significantly from the higher level of identity observed in nature for the multiple species alignment (p = 6.6×10−63).