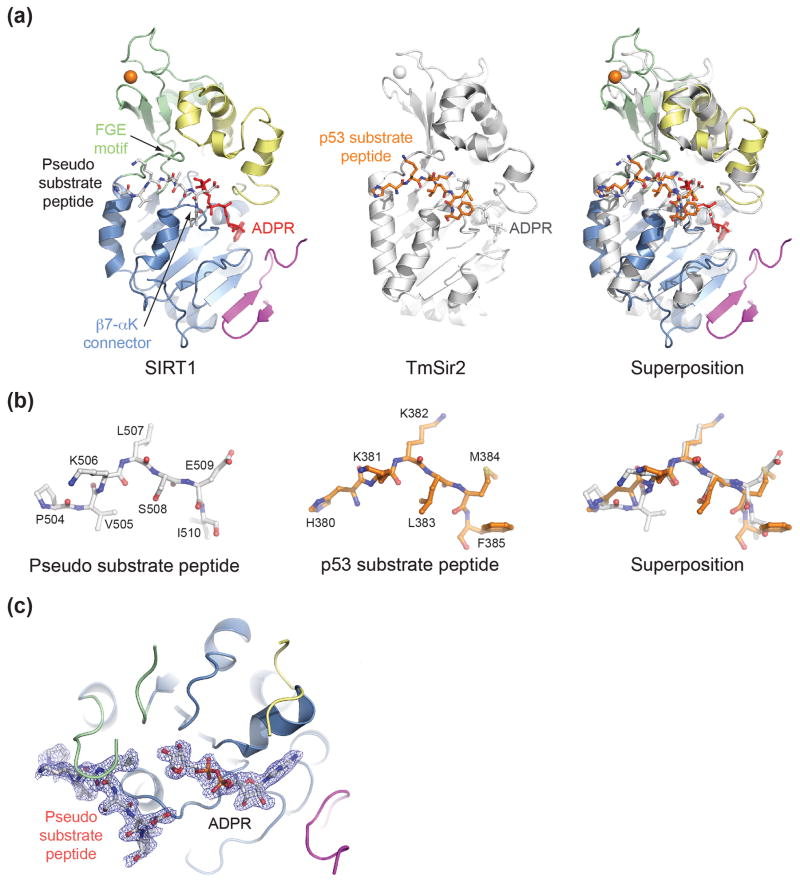
Fig. 4.
Structural comparison of H. sapiens SIRT1 to a bacterial Sir2 substrate complex. (a) A ribbon representation of SIRT1CAT•CTR•ADPR•Substrate (left, colored as in Fig. 1a), T. maritima Sir2 in complex with a p53 peptide (middle), and a superposition of the two (right). (b) A stick representation of the pseudo-substrate and p53 substrate peptide from the corresponding structures in panel (a) and their superposition. The PDB code of the TmSir2 structure is 2H59.29 (c) Final 2|fo|-|fc| electron density map around the ADPR and pseudo-substrate peptide rendered at 1.0 σ. See also Fig. S4.