Figure 2.
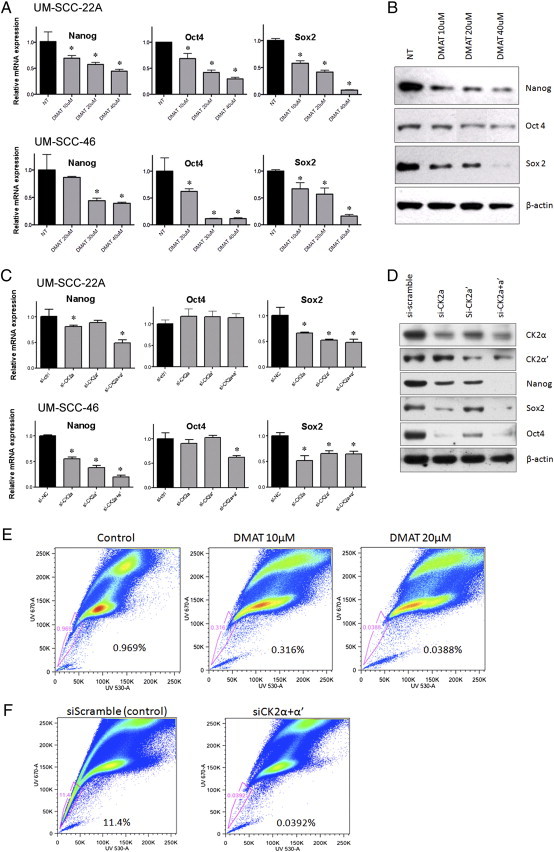
CK2 inhibition by DMAT or CK2α/α′ siRNA suppresses CSC-related SP cells and markers in two independent HNSCC lines. A. CK2 inhibitor DMAT attenuates CSC marker gene Nanog, Oct4, and Sox2 mRNA expression in UM-SCC-22A (Upper panel) and UM-SCC-46 (Lower panel). Quantitation of target genes was assessed using QRT-PCR with the treatment of increasing concentration of DMAT. Bars, SD. *Reduced versus non-treated, student t test, P < .05. B. Western blot showing Nanog, Oct4, and Sox2 protein expression are decreased in whole cell extracts from UM-SCC-46 48 hours after treatment with increasing concentrations of DMAT. β-Actin was used as loading control. Similar results for UM-SCC-22A are shown in Suppl Figure 1A. C. Effect of individual and combined CK2α and α′ siRNA knockdown on CSC marker Nanog, Oct4, and Sox2 mRNA expression in UM-SCC-22A (Upper panel) and UM-SCC-46 (Lower panel). The expression level in control siRNA-treated sample is normalized to 1.0. Columns, mean between triplicate samples; bars, SD. *Student t test, P < 0.05. D. Effect of individual and combined CK2α and α′ siRNA knockdown on CSC marker protein expression in UM-SCC-46, 48 hours after transfection with siRNA of CK2α, CK2α′, CK2α + α′, and scramble siRNA control. Nanog, Sox2, Oct4, CK2α, CK2α′ were detected, with β-actin used as loading control. Similar results for UM-SCC-22A are shown in Suppl Figure 1B. E. CK2 inhibitor DMAT and F. CK2α/α′ siRNAs inhibit SP cell detection in HNSCC. UM-SCC-46 cells pre-treated with 10 or 20 μM DMAT or DMSO diluent containing medium alone, or 100 nM scrambled control or CK2α/α′ siRNAs were labeled with Hoechst 33342 dye, and analyzed by flow cytometry. The percent HoechstLow SP cells highlighted within the window indicated were reduced by CK2 inhibitors.
