Fig. 3.
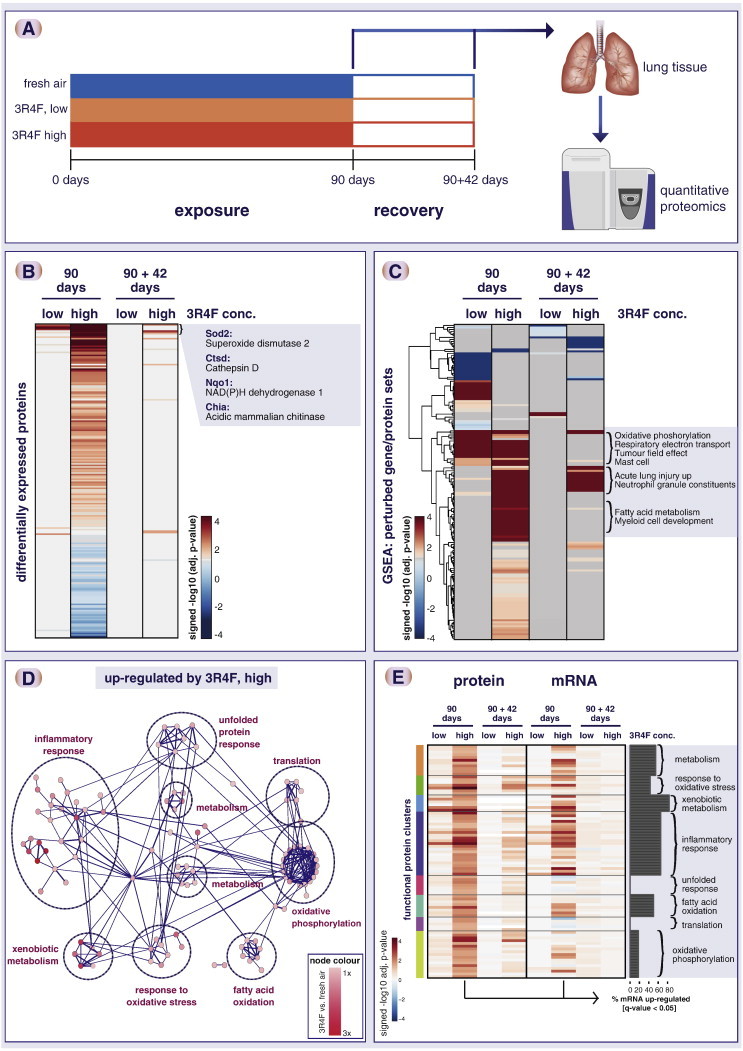
Impact of cigarette smoke exposure on the rat lung proteome. (A) Summary of rat exposure study. (B) Tobacco smoke exposure showed strong overall impact on the lung proteome. Heatmap shows significantly altered proteins (FDR-adjusted p-value < 0.05) in at least one cigarette smoke exposure condition. Each row represents a protein, each column a sample (six biological replicates), and the log2 fold-change expression values compared with sham (fresh air) exposure is color-coded. (C) Gene set enrichment analysis (GSEA) shows a concentration-dependent gene set perturbation by cigarette smoke and a partial recovery after 42 days of fresh air exposure. The heatmap shows the significance of association (− log10 adjusted p-value) of up- (red) and down- (blue) regulated proteins with gene sets. Select gene sets enriched for up-regulated proteins by cigarette smoke exposure are highlighted for three different clusters. (D) Functional interaction network of significantly up-regulated proteins upon cigarette smoke exposure shows affected functional clusters including xenobiotic metabolism, response to oxidative stress, and inflammatory response. (E) Overall, the identified functional clusters show corresponding mRNA upregulation. mRNA expression changes were measured for the same lung tissue samples and compared with the protein expression changes. The heatmap compares differential protein (left) and mRNA (right) regulation (signed − log10 q-value) for the identified protein clusters and exposure conditions. The bar plot indicates the percent of the genes that show consistent, statistically significant up-regulation of the mRNA transcript upon 90-day smoke exposure (q-value < 0.05). Note that—while overall consistent—the “translation” and “unfolded protein response” clusters show less mRNA up-regulation.
