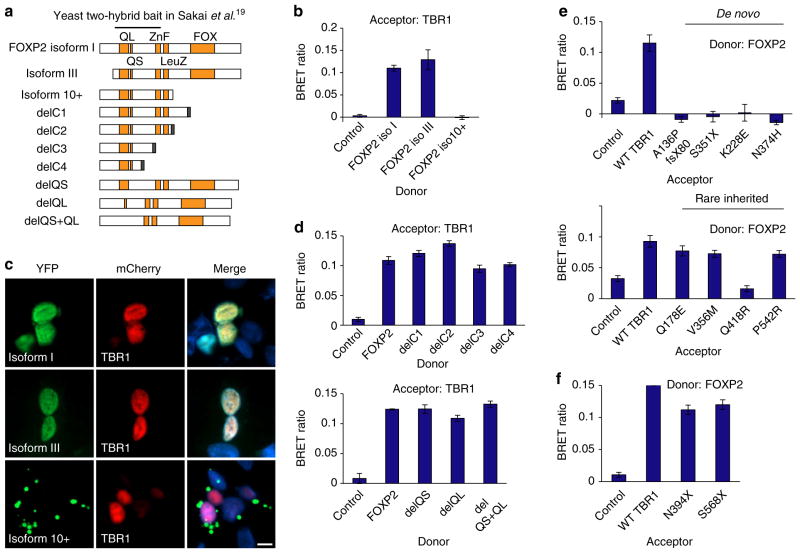
Figure 6. TBR1 interacts with FOXP2 through the T-box domain.
(a) Schematic representation of recombinant FOXP2 proteins used in BRET assays. FOXP2 contains long (QL) and short (QS) polyglutamine tracts, and zinc finger (ZnF), leucine zipper (LeuZ) and FOX DNA-binding domains. Isoforms I, III and 10 +represent natural isoforms, other constructs are synthetic. A nuclear-targeting signal appended to the C terminus in variants delC1-delC4 is indicated in black. The putative TBR1-binding region based on yeast two-hybrid data19 is also shown. (b) BRET assays for interaction between WT TBR1 and naturally occurring FOXP2 isoforms. (c) Fluorescence images of HEK293 cells co-transfected with TBR1 and FOXP2 variants. FOXP2 variants fused to YFP are shown in green (left-hand side), whereas TBR1 fused to mCherry is shown in red (middle). Nuclei were stained with Hoechst 33342 (blue). Scale bar, 5 μm. Concordant results were seen in SHSY5Y cells, as shown in Supplementary Fig. 7. (d) BRET assays for interaction between WT TBR1 and synthetic FOXP2 variants. (e) BRET assays for interaction between WT FOXP2 and TBR1 variants found in ASD. (f) BRET assays for interaction between WT FOXP2 and synthetic TBR1 truncations. In b, d–f, bars represent the corrected mean BRET ratios±s.e.m. of one experiment performed in triplicate. BRET assays were performed in HEK293 cells.