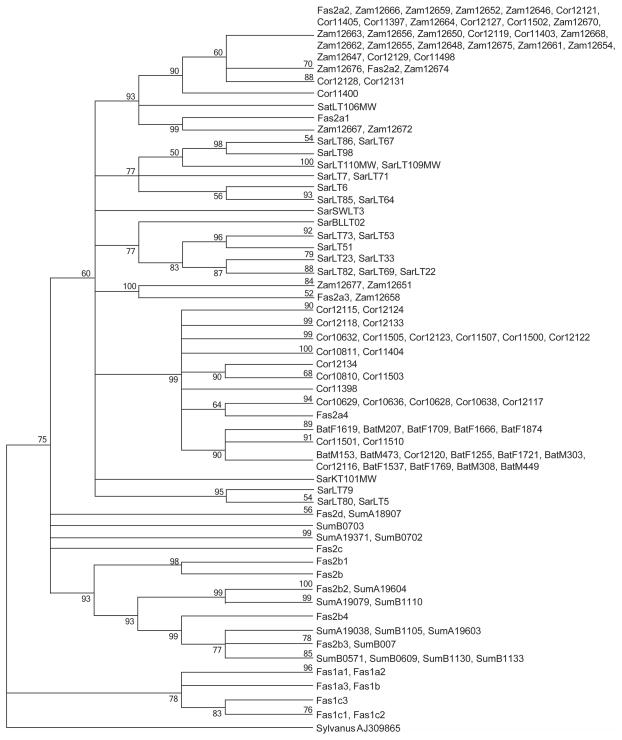
Figure 4.
Maximum Likelihood tree for a 697 bp fragment of mtDNA of five regional populations of cynomolgus macaques based on the Tamura-Nei model with 500 replicates. Branches corresponding to partitions reproduced in fewer than 50% bootstrap replicates were collapsed. Bootstrap values are provided for each branch. Sequences with “Fas” prefixes are reference sequences representing previously identified mtDNA haplogroups in cynomolgus macaques (Smith et al., 2007). Each such reference is followed by the identification number of the sample representative of that haplogoup whose prefix identifies the country of origin of that sample. Sequences with prefixes “Cor”, “Luz”, “Zam”, “Sar”, “Sum”, “Phi”, “Viet”, “Mcy”, “Maur”, and “Imcy” are sequences of samples from Corregidor, Luzon (Batangas), Zamboanga, Sarawak, Sumatra, Philippines (Mindanao), Vietnam, Malaysia, Mauritius and Indonesia, respectively. The tree is rooted with a sequence from M. sylvanus.