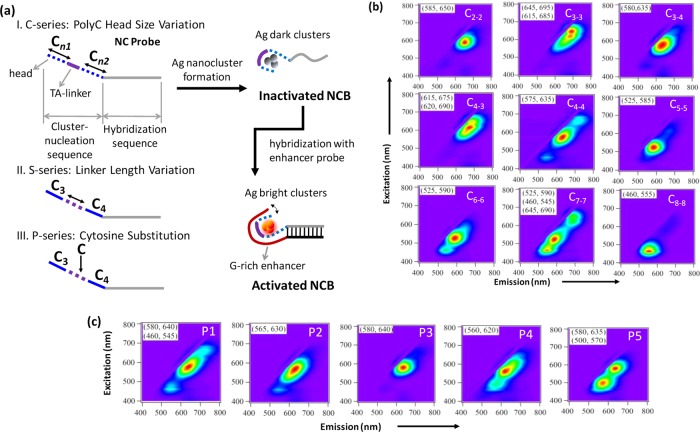
Figure 1.
(a) Schematic showing various cluster-nucleation sequence design strategies in the 3 series of experiments (C-, S-, and P-series). (I) C-series, where the size of the “polycytosine heads” is changing in the cluster-nucleation sequence; (II) S-series, where the length of the “TA-linker” is changing; and (III) P-series, where one of the linker nucleotides is replaced by a cytosine. A NanoCluster Beacon (NCB) consists of a nanocluster (NC) probe (having a C-rich cluster-nucleation sequence at the 5′-end) and an enhancer probe (having a G-rich enhancer sequence at the 3′-end). When the enhancer probe is brought close to the NC probe through hybridization, the templated dark silver clusters are activated and become highly emissive through the interactions with the nearby enhancer sequence. We call this process “the guanine-proximity-induced activation of silver clusters” or, in short, “the activation of NCBs”. (b) Normalized 2D fluorescence contour plots of the NCBs in C-series. Here C3–4 represents a cluster-nucleation sequence design with a C3 and a C4 polycytosine heads (separated by the TA-linker). (c) 2D spectra of the P-series NCBs. P3 denotes a design with the third linker nucleotide being substituted with a cytosine. The 2D spectra of the S-series NCBs are shown in Supporting Information Figure S1. For each sample, the 2D fluorescence measurement started exactly at 1 h after the addition of the enhancer probe.