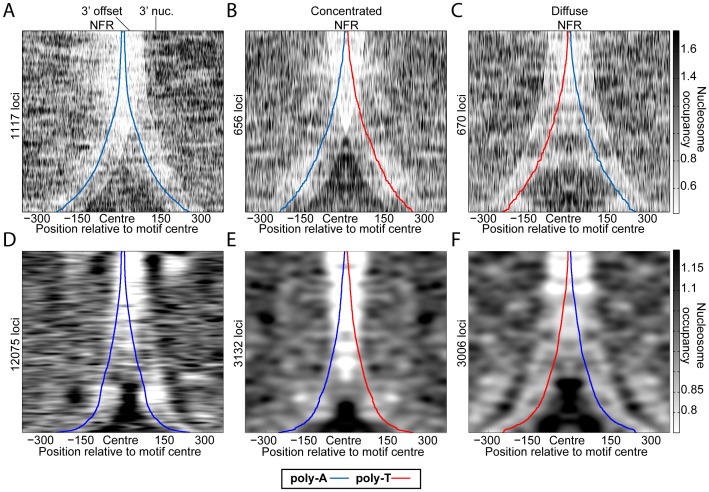
Figure 4. Mammalian nucleosome occupancy is also biased surrounding poly-As and poly-Ts, but the trend is opposite to yeast.
In vivo nucleosome occupancy for (A–C) regions with available high-resolution nucleosome data from mouse Th1 cells [9] and (D–F) non-repetitive regions on chromosome 22, for human granulocytes [10] (heatmaps) surrounding all instances of (A, D) poly-A/poly-A, (B, E) poly-A/poly-T, and (C, F) poly-T/poly-A combinations. Gaussian smoothed between rows (SD = 10 and 50, for mouse and human, respectively). The distinct transitions from light to dark in the mouse data (A-C) result from using unsmoothed data, which corresponds roughly to nucleosome dyad occupancy (in this case the poly-A/poly-T bias was more obvious without smoothing). This distinct transition is presumably caused by the destabilization of nucleosomes as poly-dA:dT tracts are incorporated, and nucleosomes appear to be most unstable when the dyad is 69 bp from the proximal poly-dA:dT tract edge in human, mouse, and yeast (Figure S4 in File S1).