Figure 1. Reads matching the human genome are more prevalent in libraries prepared from dilute samples.
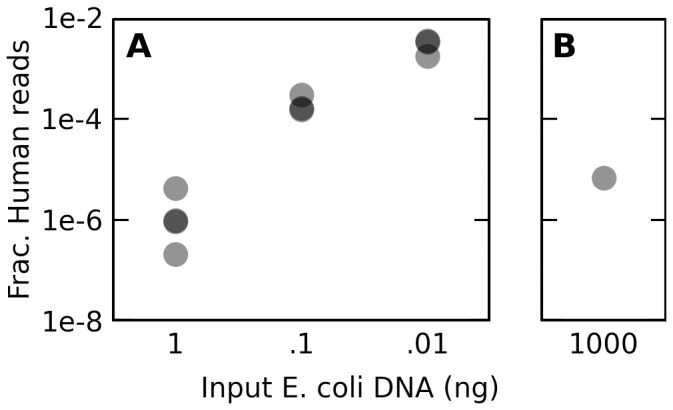
(a) The fraction of paired-end reads which preferentially map to the contaminant human genome instead of the E. coli K-12 genome, measured against the total number of reads in the library, is plotted against the amount of E. coli K-12 DNA used per tagmentation procedure as described by Parkinson et al [18]. Shading is used to highlight closely overlapping points (n = 4, 3, and 3 for the 1ng, 100pg, and 10pg libraries, respectively). Libraries listed at each concentration were not identically prepared, each using a different restriction enzyme or set of restriction enzymes at an intermediate step in the protocol (Additional File 1, Table S1), but the number and composition of enzymes used did not appreciably change the number of contaminant reads recovered. (b) The same fraction is plotted for a library prepared in the same experiment using a standard Illumina library preparation protocol. Despite a higher concentration of input DNA, an intermediate number of contaminant reads was detected.
