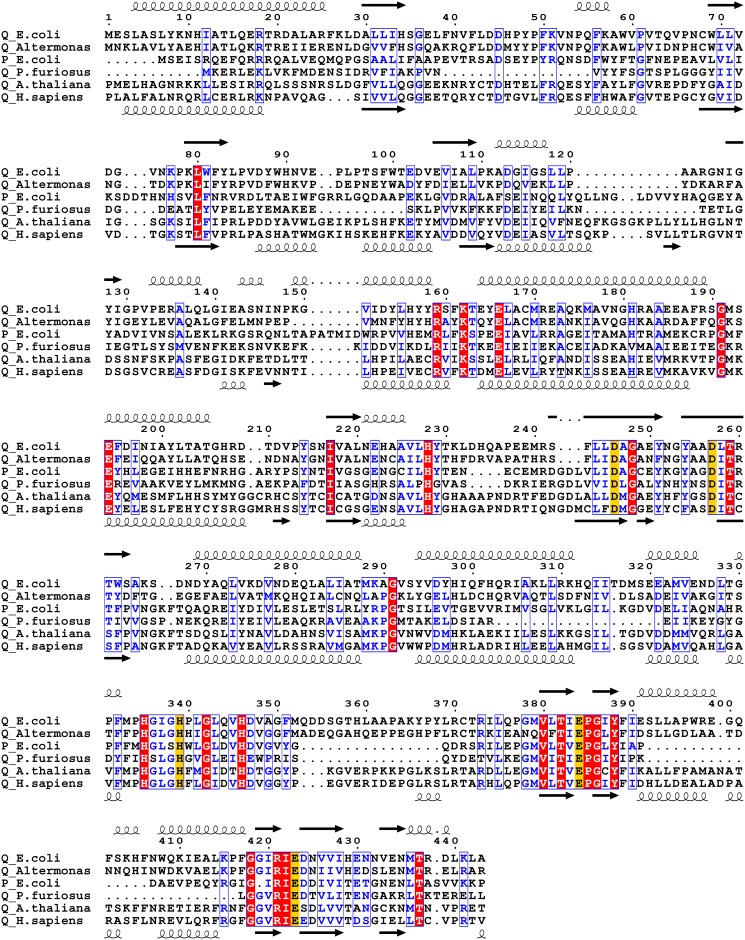
Figure 1. Sequence alignment of prolidases.
Sequence alignment of E. coli PepQ (accession number P21165) with eukaryotic and prokaryotic pita-bread fold enzymes was performed using CLUSTALW [60] and graphically organized with ESPript [61]. Completely conserved residues are highlighted in red and highly conserved residues or regions are boxed and shown in blue. Metal-chelating residues are highlighted with yellow. Numbering shown is for E. coli PepQ. Secondary structure assignments shown above the alignment are those from E. coli PepQ, while those shown below the alignment are from human PepD. The aligned proteins (with percent identity/similarity to E. coli PepQ, along with the number of aligned positions shown in parentheses; followed by the accession number of the sequence) are: Alteromonas sp. PepQ (50/67, 441), Q44238; E. coli PepP (31/46, 330), P15034; Pyrococcus furiosus PepQ (24/40, 337), P81535; Arabidopsis thaliana Xaa-Pro Dipeptidsae (34/51, 292), Q8L780; Homo sapiens PepD (29/45, 466), P12955. The degree of identity and similarity was determined by two-sequence alignment with BLAST [62].