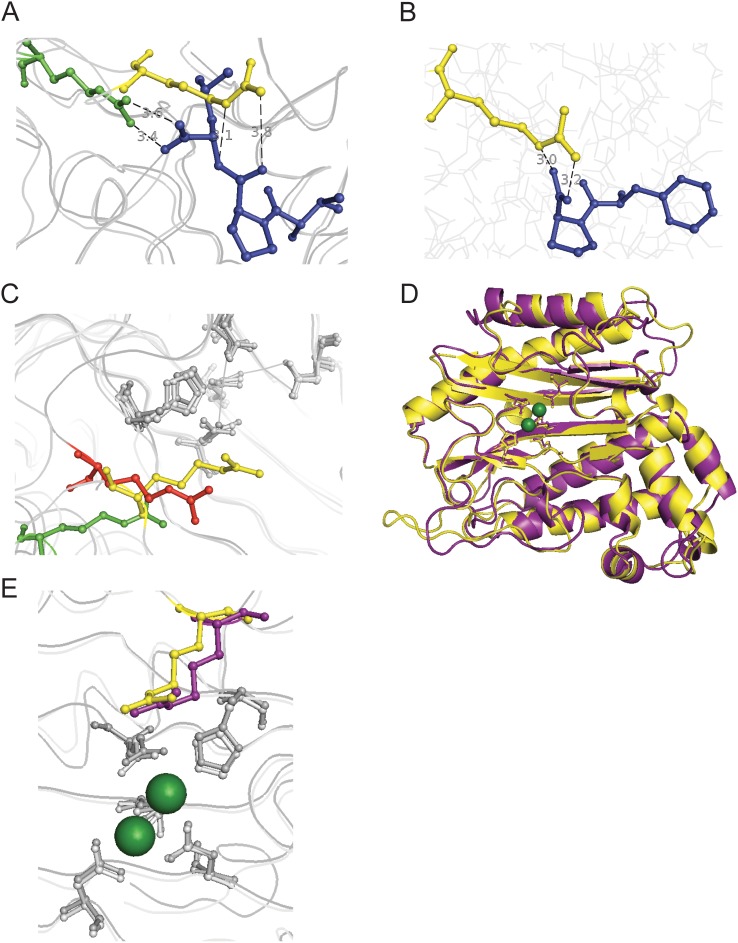
Figure 4. Structural alignment of prolidases reveals a conserved active site arginine.
(A) The PepQ catalytic domain (residues 160–443) was aligned with E. coli PepP, the proline aminopeptidase, with the bound substrate tripeptide ValProLeu (PDB entry 2BHA, residues 175–425; RMSD = 1.05 Å, 1020 atoms aligned). PepQ R370 is shown in yellow, PepP R371 is shown in green and the tripeptide is colored blue. The distances between PepP R371 and the C-terminal oxygens of the tripeptide measured at 3.4 and 3.6 Å. The distances between PepQ R370 and the prolyl-leucyl amide nitrogen and oxygen measured at 3.1 and 3.8 Å, respectively. (B) Docking simulations were performed between PepQ (yellow) and substrate dipeptides using AutoDock Vina [38]. Shown is the substrate PhePro (blue). The distances between R370 and the dipeptide C-terminal oxygens measured at 3.0 and 3.2 Å. (C) E. coli PepQ (yellow) and PepP (green) were aligned with P. furiosus prolidase (PDB entry 1PV9, residues 124–345, red). PepQ R370, PepP R371 and P. furiosus R295 are highlighted. (RMSDEcoli Q–PfuriosusQ = 0.92 Å, 816 atoms aligned; RMSDEcoli P–PfuriosusQ = 0.82 Å, 908 atoms aligned) (D) Structure alignment of catalytic domains of E. coli PepQ (yellow) and human PepD (PDB entry 2IW2, residues 187–470, purple; RMSD = 0.97 Å, 1179 atoms aligned). (E) R370 in PepQ (yellow) is sequentially and structurally conserved in humans (R398, purple). All structural alignments and distance measurements were performed with PyMOL [63].