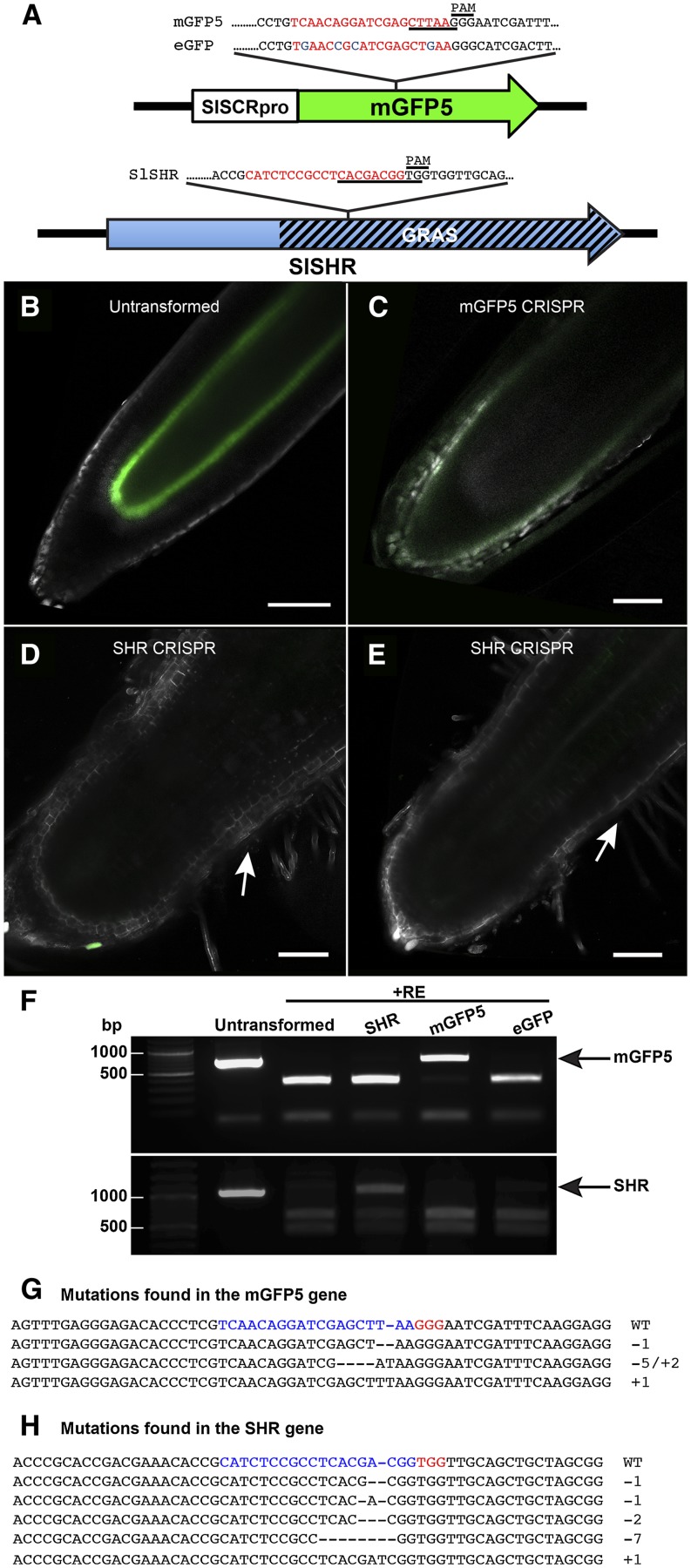
Figure 6.
The CRISPR/Cas9 system introduces mutations in hairy root transformants and can be used to study gene function in roots. A, Schematic representation of SlSCRpro-mGFP5 and SlSHR gene structures, DNA sequences, and relative locations of the targets designed for use with CRISPR. The restriction site used to screen for mutations in each target is underlined. The target sequence is in red. SNPs between mGFP5 and eGFP are in blue. The PAM follows the consensus sequence NGG. B, Control hairy root (containing no CRISPR binary vector) induced from the SlSCRpro-mGFP5 transgenic line shows strong ERGFP expression in the QC and endodermis. C to E, sgRNAs designed to complement mGFP5 and SlSHR coding sequences were transformed in an SlSCRpro-mGFP5 transgenic background using hairy root transformation. An sgRNA designed to complement mGFP5 led to the reduction or elimination of SlSCRpro-mGFP5 expression (C). Hairy root transformation with an sgRNA designed against the endogenous SHR coding sequence induced short roots with stunted meristematic and elongation zones as well as the reduction or elimination of SlSCRpro-mGFP5 expression (D and E). White arrows point to root hairs and the start of the maturation zone 0.3 mm from the root tip. This domain is outside the field of view in the control (B). B to E were imaged with a 20× objective and constant settings of 488-nm excitation, 70% laser power, 1.87-Airy unit pinhole, and 583 gain. GFP expression is shown in green with both autofluorescence spectra, and residuals are shown in white. Bars = 100 μm. F, Genomic DNA was extracted from roots transformed with Cas9 and sgRNAs against SHR, mGFP5, and eGFP. DNA was predigested with AleI and AflII for SHR and mGFP5/eGFP, respectively, and genes were amplified using primers flanking the target sequence. Amplicons were digested again. Untransformed indicates SlSCRpro-mGFP5 (same as in B) but not transformed with Cas9/CRISPR. Black arrows indicate no digestion. G and H, Alignment of sequences with Cas9-induced mutations obtained from roots transformed with Cas9/sgRNA for mGFP5 (G) and SHR (H). The wild-type (WT) sequence is shown at the top. The sequence targeted by the synthetic sgRNA is shown in red, and mutations are shown in blue. The changes in length relative to the wild type are shown to the right.