A plant-specific protein controls the complexity and connectivity of veins in cotyledons.
Abstract
The molecular mechanisms by which vascular tissues acquire their identities are largely unknown. Here, we report on the identification and characterization of VASCULATURE COMPLEXITY AND CONNECTIVITY (VCC), a member of a 15-member, plant-specific gene family in Arabidopsis (Arabidopsis thaliana) that encodes proteins of unknown function with four predicted transmembrane domains. Homozygous vcc mutants displayed cotyledon vein networks of reduced complexity and disconnected veins. Similar disconnections or gaps were observed in the provasculature of vcc embryos, indicating that defects in vein connectivity appear early in mutant embryo development. Consistently, the overexpression of VCC leads to an unusually high proportion of cotyledons with high-complexity vein networks. Neither auxin distribution nor the polar localization of the auxin efflux carrier were affected in vcc mutant embryos. Expression of VCC was detected in developing embryos and procambial, cambial, and vascular cells of cotyledons, leaves, roots, hypocotyls, and anthers. To evaluate possible genetic interactions with other genes that control vasculature patterning in embryos, we generated a double mutant for VCC and OCTOPUS (OPS). The vcc ops double mutant embryos showed a complete loss of high-complexity vascular networks in cotyledons and a drastic increase in both provascular and vascular disconnections. In addition, VCC and OPS interact physically, suggesting that VCC and OPS are part of a complex that controls cotyledon vascular complexity.
Vascular tissues, xylem and phloem, provide mechanical strength and a transport system for water, nutrients, signaling molecules, and RNA throughout the plant body. The molecular mechanisms by which these tissues acquire their identities are largely unknown. Plant vascular tissues are specified during embryonic development (Scheres et al., 1994; Mähönen et al., 2000). In Arabidopsis (Arabidopsis thaliana), the provascular stem cells or procambium are established at the early globular stage; these cells subsequently divide periclinally, giving rise to the pericycle and more procambial cells during the late globular stage of development. At the heart and torpedo stages, the number of procambium cell files in the embryo axis increases and a network of procambial strands arises in cotyledons (Scheres et al., 1994; Mähönen et al., 2000).
The architecture and spatial organization of the vascular system vary among plant organs. In Arabidopsis, the vein pattern in cotyledons is much simpler and regular than that of adult leaves (Dhondt et al., 2012). Typically, Arabidopsis cotyledons develop a midvein (or primary vein) and secondary veins that branch from the midvein and can merge to form closed areoles. During development, the embryo differentiates a complete network of procambial strands (Sieburth, 1999). After germination, the differentiation of vascular bundles into phloem and xylem is completed (Scarpella et al., 2006; Sieburth and Deyholos, 2006; Scarpella and Helariutta, 2010). This temporal separation between the specification of embryonic procambial strands and postgermination vascular differentiation offers a unique opportunity to study both processes separately. As the root and the shoot grow, new procambial cells are generated by the activity of the stem cells in the apical meristem regions. The cambium is a secondary meristem that partially derives from the procambium in adult plants and is responsible for secondary growth in the diameter of stems and roots through the production of secondary phloem and xylem. In Arabidopsis, the development of the cambium is confined to the base of the inflorescence stem, the hypocotyls, and the mature root (Zhang et al., 2011).
Over 40 genes, including different families of transcription factors, auxin carriers, auxin- and ethylene-responsive factors, Leu-rich repeat receptor-like kinases (RLKs), enzymes involved in sterol synthesis, and proteins of unknown function, have been shown to affect different aspects of vascular development in Arabidopsis (Petricka et al., 2008; Truernit et al., 2012; Miyashima et al., 2013). In addition, several plant hormones, such as cytokinins, auxin, GAs, and ethylene, have been implicated in the regulation of cambial activity (Elo et al., 2009).
Auxin plays a key role in the initiation and maintenance of procambial cells (Donner et al., 2009). Auxin canalization through the procambial cells controls the formation of continuous procambial strands and, later on, continuous vascular bundles (Sachs, 2000). Auxin canalization is achieved by the expression and polarized localization of the auxin efflux carrier PIN FORMED1 (PIN1) in procambial cells (Sauer et al., 2006; Scarpella et al., 2006). Defects in the proper polar localization of PIN1, such as those in the forked1 and scarface1 mutants, result in abnormal vein patterns and vascular discontinuities in cotyledons and leaves (Deyholos et al., 2000; Sieburth et al., 2006; Hou et al., 2010). Consistent with the role of membrane sterols in the localization of auxin transporters at the plasma membrane (Willemsen et al., 2003; Men et al., 2008; Yang et al., 2013), many sterol biosynthetic mutants show altered auxin distribution and vein pattern defects (Souter et al., 2002; Carland et al., 2010; Pullen et al., 2010).
The early expression during embryogenesis of auxin-responsive transcription factors that act as positive regulators of vascular differentiation further supports the concept that vascular cell fate decisions are made early during embryo development. For example, the auxin response factor MONOPTEROS and HOMEOBOX8 transcription factors are expressed in procambial cells at the late globular and late heart embryo stages, respectively (Baima et al., 1995; Hardtke and Berleth, 1998; Hamann et al., 1999).
Some membrane-localized receptor-mediated signaling modules have also been identified as key components in vascular specification and differentiation. Three Leu-rich repeat RLKs, RECEPTOR-LIKE PROTEIN KINASE1, TOADSTOOL2, and BRASSINOSTEROID-INSENSITIVE1-LIKE2 (BRL2), are known to be involved in vascular patterning in embryos (Nodine et al., 2007; Ceserani et al., 2009). Other RLKs, such as PHLOEM INTERCALATED WITH XYLEM (Fisher and Turner, 2007; Etchells and Turner, 2010; Etchells et al., 2013), XYLEM INTERMIXED WITH PHLOEM1 (Bryan et al., 2012), MORE LATERAL GROWTH1, and REDUCED IN LATERAL GROWTH1 (Agusti et al., 2011), control vasculature identity and development in adult tissues, but their roles in embryonic vasculature patterning have not been analyzed.
Recently, the plant-specific, membrane-associated protein OCTOPUS (OPS) was shown to be important in vascular development, although its molecular function remains unknown (Truernit et al., 2012).
Understanding the mechanisms underlying plant vascular development is not only important from a biological perspective but also from an economic point of view. A major fraction of the plant biomass that can be used as a source of renewable energy through conversion into biofuels comes from vascular tissues. Increase in vascular complexity in stems has been associated with an increase in tissue density in Arabidopsis (Ibañes et al., 2009). In addition, the activity levels of the procambium and vascular cambium and the general increase in the number of cells differentiating secondary cell walls such as xylem cells have dramatic effects in biomass yield in dicots (Hejátko et al., 2009; Etchells and Turner, 2010; Wang et al., 2010).
In this article, we analyze the function of At2g32280, a gene coding for a Domain of Unknown Function1218 (DUF1218)-containing protein with four predicted transmembrane domains. Arabidopsis knockout mutants for At2g32280 show abnormal vascular networks in cotyledons, leading us to name this gene VASCULATURE COMPLEXITY AND CONNECTIVITY (VCC). VCC belongs to a 15-member gene family and is required for proper embryo provasculature development.
RESULTS
DUF1218-Containing Proteins in Arabidopsis and Other Plants
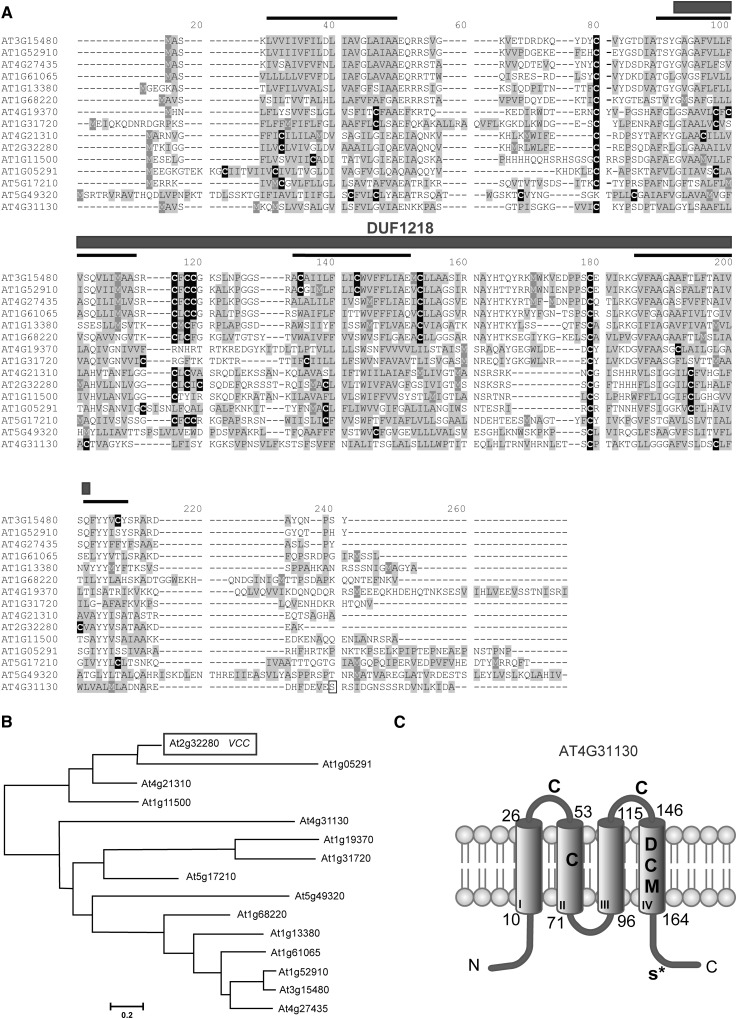
Some genes encoding DUF1218-containing proteins have been shown to be differentially expressed during the in vitro differentiation of tracheary elements (Kubo et al., 2005). In addition, a DUF1218-containing protein (AT2G68220) interacts with the RLK BRL2 that is involved in vascular patterning (Ceserani et al., 2009). To explore a putative role of DUF1218-containing proteins in vascular development, we searched in the Arabidopsis genome and found that it contains 15 genes coding for proteins with four predicted transmembrane regions and a DUF1218 domain (Fig. 1, A and B). The DUF1218 domain consists of approximately 100 amino acids with several conserved Cys residues (Fig. 1A). To analyze the putative evolutionary history of this domain, we searched for DUF1218-containing protein sequences from different organisms and performed a phylogenetic analysis. We were able to identify DUF1218-containing protein sequences only in land plants, from liverworts to flowering plants. No DUF1218 homologous sequences were found in algae or in nonplant organisms, suggesting that DUF1218 is a plant-specific domain. Mining the databases at Phytozome (http://www.phytozome.net/), The Arabidopsis Information Resource (http://www.arabidopsis.org/), and the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), we obtained 63 sequences containing DUF1218 domains from Arabidopsis (15 sequences), Medicago truncatula (11 sequences) Brachypodium distachyon (16 sequences), Physcomitrella patens (13 sequences), and Marchantia polymorpha (two EST sequences from the NCBI). We aligned these 63 sequences with ClustalW and performed a phylogenetic analysis using maximum likelihood with MEGA5 (Tamura et al., 2011; Supplemental Fig. S1). Some genes within this family, such as At4g31130 and At5g17210, seem to have diverged very early during the evolution of land plants and have orthologs in mosses, ferns, and other flowering plants. However, the majority of the Arabidopsis DUF1218 proteins grouped into clades containing only protein sequences from flowering plants, suggesting a more recent diversification (Supplemental Fig. S1).
Figure 1.
DUF1218-containing proteins. A, Alignment of Arabidopsis DUF1218-containing proteins. Predicted transmembrane domains are indicated by a black line on top of the sequence; Cys residues (black boxes) and hydrophobic residues (gray boxes) are highlighted. The phosphorylated Ser (Ser-177) identified by Hem et al. (2007) in AT4G31130 is indicated by a white box. B, Distance tree of DUF1218-containing proteins in Arabidopsis generated with the MEGA5 software. C, Transmembrane domains (I–IV) and protein topology for AT4G31130 predicted by TMpred (Hofmann and Stoffel, 1993). The DUF1218 domain includes transmembrane domains II to IV. Asp (D) and Met (M) residues in the fourth transmembrane domain, Cys (C) residues in both transmembrane and extracellular domains, and the phosphorylated Ser-177 (S*) are indicated.
In Arabidopsis, DUF1218-containing proteins range from 163 to 257 amino acids, with no other predicted conserved domains. According to the TMpred software (Hofmann and Stoffel 1993; http://www.ch.embnet.org/software/TMPRED_form.html), DUF1218-containing proteins are predicted to have four transmembrane domains, three of which are contained within the DUF1218 region (Fig. 1, A and C). A proteomic analysis of Arabidopsis plasma membrane proteins identified a phosphorylated residue (Ser-177) close to the C terminus of the DUF1218-containing protein encoded by At4g31130 (Hem et al., 2007), suggesting that the N- and C-terminal regions plus the central loop of this protein are in the cytoplasm and the first and third loop are exposed to the lumenal/extracellular space (Fig. 1C). Several Cys residues (black boxes in Fig. 1A) in the transmembrane domains and in the luminal/extracellular loops are well conserved across the whole DUF1218 protein family. In addition, several polar amino acid residues are found along the transmembrane domains (Fig. 1, A and C).
VCC Regulates Vascular Network Complexity and Connectivity in Cotyledons
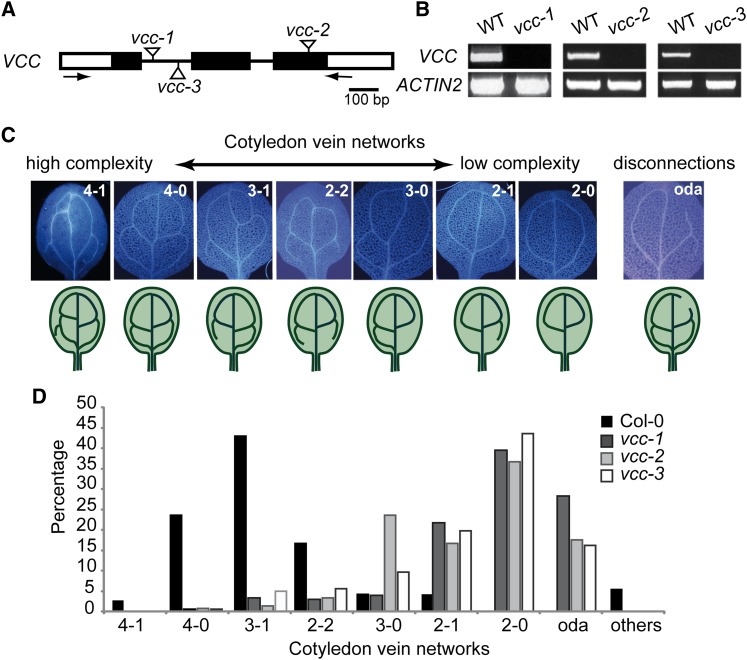
To analyze the function of DUF1218-containing proteins in Arabidopsis, we obtained and characterized transfer DNA (T-DNA) insertional mutants for members of this gene family. Three independent mutant lines for At2g32280 showed abnormal vein networks in cotyledons of 1-week-old seedlings (Fig. 2), and thus we named this gene VCC. We designated these mutant alleles vcc-1, vcc-2, and vcc-3. VCC gene organization and the position of the T-DNA insertions are shown in Figure 2A. Reverse transcription (RT)-PCR analysis of RNA extracts from mutant plants demonstrated that vcc-1, vcc-2, and vcc-3 are knockout mutants, as they lack detectable VCC transcripts (Fig. 2B).
Figure 2.
Mutant vcc seedlings show abnormal cotyledon vein networks. A, VCC genomic organization. Black boxes represent exons, and white boxes represent untranslated regions. T-DNA insertion sites for the vcc-1, vcc-2, and vcc-3 alleles are indicated. Arrows indicate the locations of primers used in RT-PCR analysis. B, RT-PCR analysis of VCC expression in control (wild-type [WT]) and mutant vcc plants. Amplification of ACTIN2 mRNA was used as a control. C, Classification of vein complexity patterns in cotyledons from 7-d-old seedlings based on the number of closed areoles (two, three, or four) formed by the secondary veins and the number of vein branches/incomplete areoles in the proximal (closest to the petiole) part of the cotyledon. Open distal areoles (oda) were classified as vein disconnections. D, Distribution of vein complexity patters in wild-type Col-0 and vcc-1, vcc-2, and vcc-3 mutants (n = 72 for wild-type Col-0, 304 for vcc-1, 365 for vcc-2, and 221 for vcc-3). [See online article for color version of this figure.]
To analyze changes in cotyledon vascular organization in vcc lines, we classified patterns of vein complexity into different categories (Fig. 2C). We considered the number of closed areoles (two, three, or four) formed by the secondary veins and the number of vein branches/incomplete areoles in the proximal (closest to the petiole) part of the cotyledon. Open distal (closest to the apex) areoles resulted from vein gaps and were considered vein disconnections. Whereas more than 60% of wild-type cotyledons showed either four closed areoles (4-0) or three closed areoles with one open proximal areole (3-1; Fig. 2D), approximately 40% of the cotyledons from the three independent vcc mutant lines showed a simpler vein network with only two closed areoles (2-0). In addition, in contrast to wild-type cotyledons in which the distal areoles are almost always closed (99% of the examined wild-type cotyledons), cotyledons of the three vcc mutant lines displayed between 17% (vcc-2) and 28% (vcc-1) open distal areoles (Fig. 2D; Supplemental Table S1). This indicates that mutations in VCC negatively affect the complexity and connectivity of cotyledon veins.
No defects in vascular patterning or differentiation were detected in any other organs or developmental stage in vcc mutants.
Overexpression of VCC Induces Changes in Vein Complexity in Cotyledons and Leaves
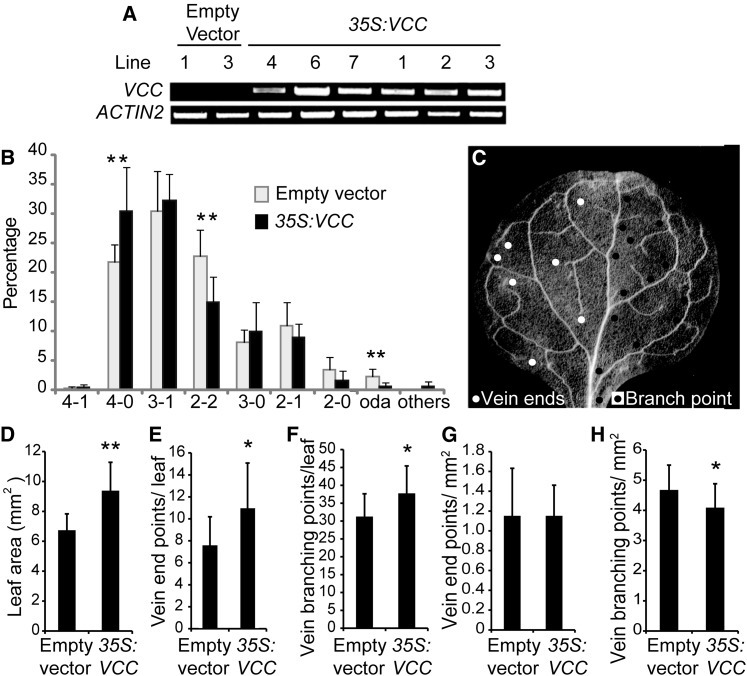
To analyze the effect of overexpressing VCC in plants, we transformed wild-type plants with a construct containing pCaMV35S:VCC or with an empty vector as a control. We isolated multiple T1 transgenic lines and determined VCC mRNA accumulation by RT-PCR (Fig. 3A). We then analyzed vascular network organization in cotyledons of T2 seeds from seven control lines transformed with the empty vector and nine pCaMV35S:VCC plants (Fig. 3B; Supplemental Table S2). The VCC-overexpressing lines showed an increase in cotyledon vein complexity, as evidenced by a 10% increase in cotyledons with four closed areoles (4-0) and a 10% decrease in cotyledons with two closed distal areoles and two open proximal areoles (2-2). For six of the nine VCC-overexpressing lines, between 30% and 45% of cotyledons showed four closed areoles (4-0 and 4-1), whereas no control line showed more than 26% of cotyledons with these categories (Supplemental Table S1).
Figure 3.
VCC overexpression increases the complexity of vein networks in cotyledons and first leaves. A, RT-PCR analysis of VCC expression in Col-0 plants transformed with either an empty vector or the pCaMV35S:VCC construct (35S:VCC). cDNA was obtained using 1 μg of RNA, and 2 μL of RT product was used as a template in a 25-μL volume reaction. Twenty-five PCR cycles were performed to amplify VCC. ACTIN2 was used as a control. B, Distribution of vein complexity patterns in pCaMV35S:VCC and plants transformed with an empty vector. T2 embryos from seven T1 lines transformed with an empty vector and nine T1 pCaMV35S:VCC lines were analyzed. For each line, between 100 and 200 individual cotyledons were examined. C, Dark-field image of a first rosette leaf from a 3-week-old Arabidopsis plant. For clarity, vein end points (white circles at left side) and vein branching points (black circles at right side) considered for vein complexity analysis are indicated in only half of the leaf blade. D, Quantitative analysis of first leaf area. E, Quantification of vein end points and branching points per leaf. F, Vein branching points per leaf. G, Ratio between vein end points and leaf blade area. H, Ratio between vein branching points and leaf blade area. In total, 12 and 28 first rosette leaves from T1 plants transformed with the empty vector and the pCaMV35S:VCC construct, respectively, were analyzed. For Student’s t test analysis, P < 0.05 (*) and P < 0.01 (**).
To determine whether VCC overexpression can also affect vasculature organization in rosette leaves, we analyzed the first leaves of 3-week-old T1 plants. To evaluate leaf vein complexity patterns, we quantified the number of vein branching points and free-ending veins according to Dhondt et al. (2012; Fig. 3C). We found that, compared with control plants transformed with an empty plasmid, the VCC-overexpressing leaves were significantly larger in surface area (Fig. 3D), also showing more vein ends and vein branching points per leaf (Fig. 3, E and F). Whereas no differences were found in the number of vein end points per leaf area, the number of vein branching points per leaf area was higher in VCC-overexpressing leaves compared with control leaves (Fig. 3, G and H), suggesting that the overexpression of VCC partially affects the complexity of the leaf vein networks. We did not detect alterations in vascular organization in any other organ of VCC-overexpressing plants.
Provascular Tissue Differentiation in vcc Cotyledons
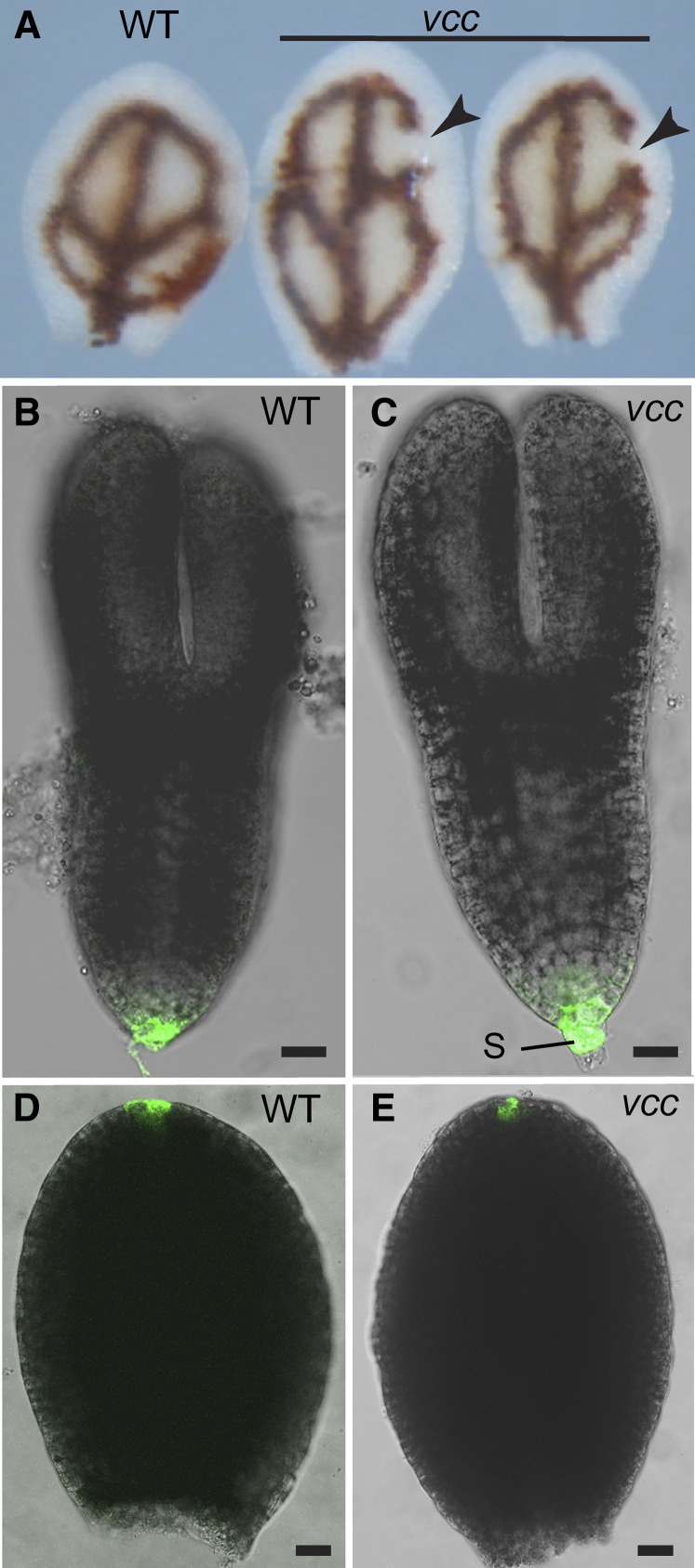
The alterations in vascular complexity in cotyledon observed in vcc mutants could be explained by at least two mechanisms. Mutations in VCC could affect either the patterning of procambial cell specification in cotyledons or the differentiation of vascular tissues from normally established procambial strands. If the first scenario is correct, cotyledon procambial strands should show the same architectural alterations seen in differentiated vascular bundles after germination. To determine the distribution of procambial strands, we analyzed iron accumulation patterns in developing embryos. Iron accumulates inside vacuoles of the proendodermal layer around procambial strands during embryo maturation; iron detection has been used successfully as a marker to visualize defects in provasculature continuity (Roschzttardtz et al., 2009, 2010). The type and frequency of proendodermal/procambial networks revealed by iron staining in vcc mutant developing embryos (Fig. 4A; Supplemental Fig. S2) were comparable to those observed in cotyledons from 1-week-old vcc seedlings (Fig. 2D). These results indicate that alterations in vasculature organization in the vcc cotyledons happened early during embryo provasculature development.
Figure 4.
Analysis of vcc embryos. A, Mature wild-type (WT) and vcc-2 cotyledons were stained with Perls/DAB (Roschzttardtz et al., 2009) to detect iron accumulation in the proendodermis that surrounds procambial strands. Vein disconnections or gaps (arrowheads) were evident in the distal areoles of vcc cotyledons. B to E, Representative examples of GFP expression patterning in wild-type Col-0 and vcc-2 embryos expressing pDR5:GFP. B and C, Torpedo embryo stage. D and E, Dissected cotyledons from bent cotyledon stage embryos. S, Suspensor. Bars = 20 μm.
Discontinuous veins are common in cotyledons of mutants with defects in either auxin response or distribution (Hobbie et al., 2000; Carland et al., 2010). To test whether auxin distribution is altered in the vcc mutant cotyledons, we obtained homozygous vcc-2 plants expressing GFP under the control of the auxin-responsive DR5rev promoter (Friml et al., 2003). We did not detect changes in DR5rev promoter activity caused by the vcc-2 mutation (Fig. 4, B–E). We also analyzed lines expressing pPIN1:PIN1-GFP and did not detect any changes in either the expression or the polar localization of PIN1-GFP in vcc-2 embryos (Supplemental Fig. S3). These results suggest that the defects in cotyledon vasculature organization caused by mutations in VCC are not associated with detectable changes in PIN1 localization or auxin distribution.
VCC Expression Pattern
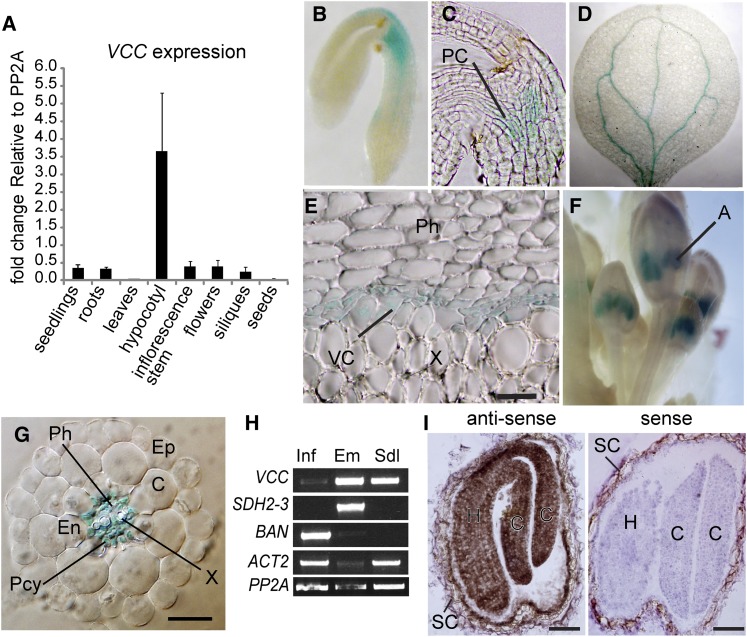
According to public microarray data, VCC mRNA seems to be more abundant in roots (phloem), hypocotyl, shoot apex, flowers, and embryos during seed development (eFP Browser; http://bar.utoronto.ca; Supplemental Figs. S4 and S5). To corroborate these data, we performed quantitative RT-PCR analysis using RNA from different tissues/developmental stages. Our results confirm that the VCC transcript is most abundant in 2-month-old hypocotyls, although it is detected in other organs/developmental stages (Fig. 5A).
Figure 5.
VCC expression pattern. A, Quantitative RT-PCR analysis of VCC transcripts extracted from rosette leaves, inflorescence stems, maturing green siliques, dry seeds, and roots. The expression of VCC was compared with PP2A. B to G, Histochemical staining to detect GUS activity in different tissues and organs of Col-0 plants transformed with pVCC:GUS. GUS activity was detected in procambial cells at the hypocotyl of mature embryos (B and C), cotyledon veins from germinated seedlings (D), the vascular cambium at the root-hypocotyl junction with secondary growth (E), anthers in floral buds (F), and cells within the root vascular cylinder, including phloem, xylem, and pericycle cells (G). H, RT-PCR of VCC transcripts from inflorescences bearing floral buds, open flowers, and developing siliques (Inf), torpedo embryos (Em), and 10-d-old seedlings (Sdl). To test for tissue contamination, transcripts of SDH2-3, which is specifically expressed in maturing embryos and dry seeds, and BAN, which is expressed exclusively in the seed coat, were also amplified. ACTIN2 (ACT2) and PP2A were used as loading controls. I, In situ hybridization of VCC transcripts on sections of developing seeds. VCC transcripts were detected using a specific antisense biotin-labeled probe, streptavidin-horseradish peroxidase, and metal-enhanced DAB. Positive detection of VCC transcripts is revealed by brown precipitates. No signal was detected with a sense biotin-labeled RNA probe (negative control). A, Anther; C, cortex; Ct, cotyledon; En, endodermis; Ep, epidermis; H, hypocotyl; PC, procambium; Pcy, pericycle; Ph, phloem; SC, seed coat; VC, vascular cambium; X, xylem. Bars = 100 μm (E), 20 μm (G), and 50 μm (I).
To analyze in more detail the expression pattern of VCC, we obtained transgenic plants expressing GUS under the transcriptional control of a 1,900-bp DNA fragment derived from a sequence located immediately upstream of the VCC coding region. Twenty pVCC:GUS transgenic lines were isolated and analyzed. GUS activity was detected in procambial cells at the hypocotyl of mature embryos (Fig. 5, B and C), in cotyledon veins after germination (Fig. 5D), in the vascular cambium at the root-hypocotyl junction of 2-month-old plants (Fig. 5E), in anthers of young flowers (Fig. 5F), and in cells within the root vascular cylinder, including pericycle, xylem, and phloem cells (Fig. 5G).
Since we failed to detect consistent GUS expression in developing embryos, we performed RT-PCR and in situ hybridization of VCC transcripts and confirmed that VCC is expressed in torpedo and bent cotyledon stage embryos (Fig. 5, H and I). The detection of VCC transcripts by in situ hybridization showed that VCC is expressed in all embryo cells, including procambial cells, at the bent cotyledon embryo stage (Fig. 5I).
We also obtained pVCC:VCC-GFP transgenic lines, but the expression of the pVCC:VCC-GFP construct did not rescue the cotyledon vein pattern defects in vcc plants, suggesting that the VCC-GFP fusion protein may be nonfunctional. Therefore, we could not use these lines to characterize the subcellular localization of VCC-GFP.
Double Mutant in VCC and OPS Shows Severe Defects in Cotyledon Veins
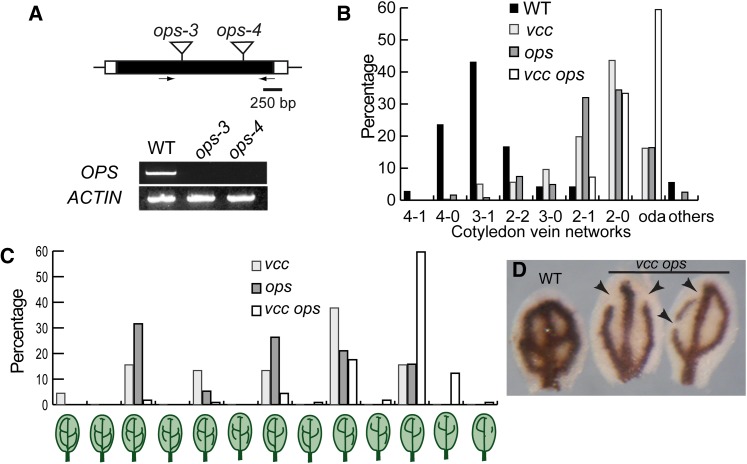
Truernit et al. (2012) reported that mutations in OPS, a membrane-associated protein expressed in procambial cells and phloem, lead to a reduction in cotyledon vascular pattern complexity and discontinuous veins in Arabidopsis. In addition, similar to VCC, the overexpression of OPS resulted in an increase in cotyledon vascular pattern complexity. Unlike VCC, OPS is not predicted to contain transmembrane domains but is assumed to associate with membranes through posttranslational lipidation (Benschop et al., 2007; Truernit et al., 2012). Because mutations and overexpression of OPS and VCC have comparable effects in the cotyledon vascular network, we tested a possible genetic interaction between VCC and OPS by analyzing double knockout vcc ops mutants. We isolated two new ops mutant alleles, ops-3 (SALK_089722C) and ops-4 (SALK_042563), and confirmed by RT-PCR analysis that the expression of OPS mRNA is undetectable in homozygous ops-3 and ops-4 T-DNA knockout mutant lines (Fig. 6A). Consistent with the published analysis of ops-1 and ops-2 (Truernit et al., 2012), ops-3 and ops-4 showed reductions in the complexity of vascular networks in cotyledons (Fig. 6B; Supplemental Table S2). Interestingly, the cotyledons from vcc-3 ops-4 double homozygous mutants showed drastic reductions in vein complexity. High-complexity vein networks (4-1, 4-0, and 3-1) were present in more than 75% of wild-type cotyledons but in less than 5% of the vcc-3 ops-4 mutant cotyledons. In addition, 60% to 70% of the vcc-3 ops-4 cotyledons showed gaps/disconnections in the distal veins (open distal areoles), which represents a 2.5- to 4-fold increase in the occurrence of vein disconnections compared with the single vcc and ops mutant lines (Figs. 2D and 6B). We also classified all the vascular networks with open distal areoles observed in vcc-3, ops-4, and vcc-3 ops-4 cotyledons (Fig. 6C). We observed that approximately 16% of the vcc ops double mutants show vascular networks with two open distal areoles per cotyledon and discontinuous/fragmented veins. These vein networks were not seen in either single ops or vcc mutants, indicating that, besides the increase in open distal areole frequency, new abnormal vascular networks appeared in the double vcc ops mutant.
Figure 6.
The vcc ops double mutant shows severe defects in the organization of cotyledon vein networks. A, OPS genomic organization and characterization of OPS mRNA expression in ops-3 and ops-4. Black boxes represent exons and white boxes represent untranslated regions. Insertion sites for the ops-3 and ops-4 alleles are indicated. Arrows indicate the locations of primers used in RT-PCR analysis. Amplification of ACTIN2 was used as a control. WT, Wild type. B, Frequency of vein complexity patterns in ops-3 (n = 82), ops-4 (n = 122), and vcc-3 ops-4 (n = 213). oda, Open distal areoles. C, Frequency of vein networks with distal open areoles and fragmented veins in cotyledons of vcc-3 (n = 44), ops-4 (n = 19), and vcc-3 ops-4 (n = 114). D, Mature wild-type and vcc-3 ops-4 cotyledons stained with Perls/DAB (Roschzttardtz et al., 2009) to reveal iron accumulation in the proendodermis that surrounds procambial strands. Multiple vein disconnections/gaps (arrowheads) were evident in the distal areoles of the mutant cotyledons. [See online article for color version of this figure.]
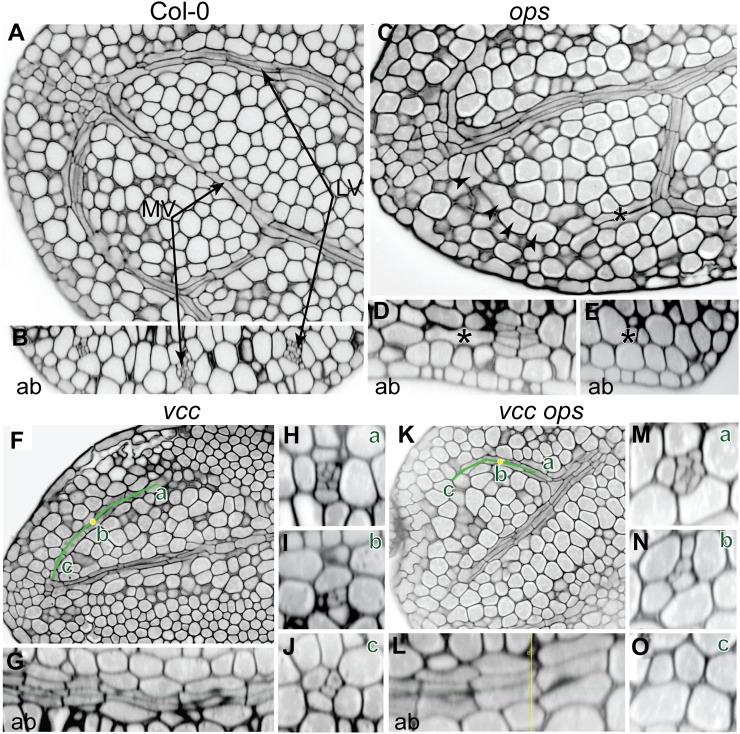
Similar to vcc mutants, embryonic iron distribution was affected in vcc-3 ops-4 cotyledons, indicating an early provasculature developmental defect (Fig. 6D). To further understand the developmental problems associated with the abnormal embryo vasculature in the vcc-3 ops-4 mutant, we stained mature embryos that were rehydrated overnight and removed from the seed coat with modified pseudo-Schiff propidium iodide (Fig. 7). The Columbia-0 (Col-0) cotyledons showed closed areoles with well-developed vascular (xylem and phloem) initials (Fig. 7, A and B). In the vcc mutant, consistent with the iron staining results (Fig. 4A), the differentiation of initials within the cotyledon veins was incomplete; we detected fewer vascular initials in the distal part of the veins, suggesting that procambial cells in the vcc mutant failed to divide to give rise to phloem and xylem cells (Fig. 7, F–J). In ops mutant cotyledons, vein gaps were due to a gradual loss of procambial cell division that led ultimately to a complete absence of procambium at the distal part of the vein (Fig. 7, C–E). Similar defects in the division of procambial cells were observed in vcc-3 ops-4 mutants but with a higher frequency, suggesting an additive effect of these two mutations.
Figure 7.
Histological characterization of vasculature in cotyledons. A and B, Median longitudinal optical sectioning of a Col-0 cotyledon (A) and its corresponding transversal sectioning (B) showing complete division of the procambium into phloem and xylem initials. C, Median longitudinal section of an ops-4 cotyledon showing a gap in the vascular networks (arrowheads). D and E, Trans-section (D) and lateral section (E) in the distal loop in the positions indicated by the asterisks showing a progressive loss of procambial divisions. F, Median longitudinal sectioning of a vcc-3 cotyledon. The green line indicates the vascular loop. G, Curved lateral section of the corresponding loop indicated by the green line in G. H to J, Three different trans-sections at different positions of the green line, from proximal (a) to median (b; yellow dot) to distal (c) positions. K, Median longitudinal section of a vcc-3 ops-4 cotyledon. L, Curved lateral section of the corresponding loop through the green line in K. M to O, Three different trans-sections at different positions of the green line, from proximal (a) to median (b; yellow dot) to distal (c) positions. Ab, Abaxial; LV, lateral vein; MV, middle vein. Bars = 10 μm.
VCC Interacts with OPS
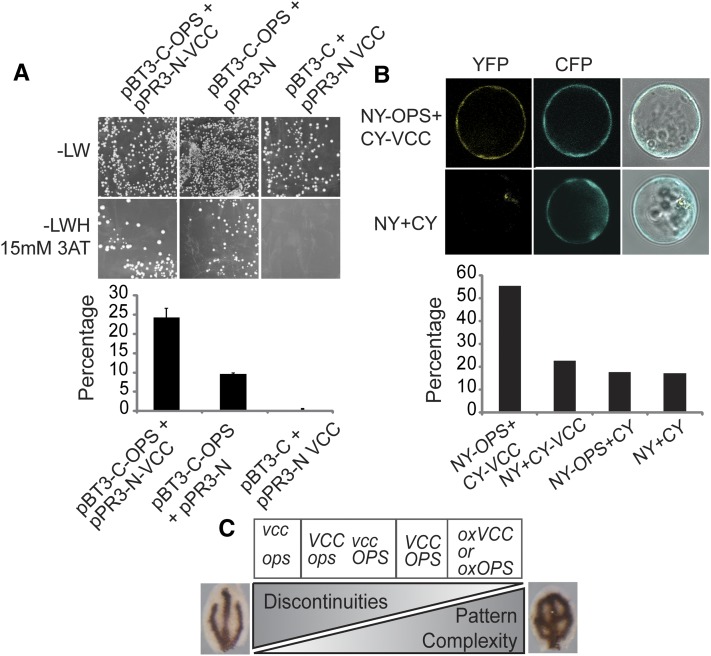
To test whether the VCC and OPS interact physically, we performed a directed split-ubiquitin yeast (Saccharomyces cerevisiae) two-hybrid assay. We cotransformed yeast cells with pBT3-C-OPS and pPR3-N-VCC, pBT3-C and pPR3-N VCC, or pBT3-C-OPS and pPR3-N and determined the number of transformant colonies grown on plates with selection medium for interaction (-Leu, Trp, His [-LWH] plus 15 mm 3-aminotriazole [3-AT]) and colonies grown on plates with section medium for plasmid transformation (-Leu, Trp [-LW]) to calculate a ratio (Fig. 8A). The coexpression of VCC as prey and OPS as bait resulted in 24% colony growth under selection for interaction, whereas we detected 0% and 9% colony growth when VCC and OPS, respectively, were coexpressed with empty vector OPS (Fig. 8A).
Figure 8.
Interaction between OPS and VCC. A, Split-ubiquitin yeast two-hybrid assay between OPS and VCC. The number of colonies grown on -LWH medium containing 15 mm 3-AT (selection for interaction) was divided by the number of colonies grown on -LW medium (selection for transformation) to calculate the percentage of colonies showing a VCC-OPS positive interaction. Controls were performed by coexpressing VCC and OPS with the corresponding prey or bait empty vectors. Between 500 and 3,000 colonies were counted in each case. The graph shows combined results from two independent experiments; error bars indicate sd. B, Bimolecular fluorescence complementation assay on Arabidopsis protoplasts. A vector containing a pCaMV35S:CFP reporter was cotransfected to assess transfection efficiency. The graph shows the percentage of CFP-positive protoplasts with YFP signal. Between 250 and 400 protoplasts were scored for each combination of vectors. C, VCC and OPS in the control of cotyledon vein patterning. Expression levels of VCC and OPS affect both pattern complexity and the connectivity of veins in cotyledons.
In addition, we tested interaction between VCC and OPS by a bimolecular fluorescence complementation assay. We coexpressed VCC fused to the C-terminal portion of yellow fluorescent protein (YFP; CY-VCC) and OPS fused to the N terminus of YFP (NY-OPS) in Arabidopsis protoplasts (Fig. 8B). As negative controls, we coexpressed each of the fusion proteins with the corresponding empty vector and the two empty vectors together (Fig. 8B). Only the coexpression of CY-VCC and NY-OPS allowed for the reconstitution of YFP fluorescence above background levels (Fig. 8B), indicating that the two proteins are able to interact in Arabidopsis.
DISCUSSION
According to the Pfam database (http://pfam.sanger.ac.uk/family/duf1218) and our own analysis of the Arabidopsis DUF1218-containing proteins, DUF1218 contains a number of conserved Cys residues and polar amino acids in the predicted transmembrane domain regions. The presence of conserved Cys residues and polar amino acids in transmembrane domains is also a distinctive feature of other proteins of known molecular function. For example, DUF1218-containing proteins share some structural features with metal transporters such as human COPPER TRANSPORTER1 (hCTR1). hCTR1 contains 190 amino acids and three transmembrane domains with polar amino acid residues that are important for metal transport (Eisses and Kaplan, 2005; Nose et al., 2006). VCC proteins also share similarities with TETRASPANIN (TET) proteins, which consist of 200 to 350 amino acids and contain several conserved Cys residues and four transmembrane domains with polar amino acid residues (Hemler, 2005). In animal cells, TETs interact with each other and with other membrane proteins, including receptors, to form TET-enriched microdomains involved in cell signaling. Different from the DUF1218 family, canonical TET proteins have a large structured second extracellular loop (Stipp et al., 2003). However, this extracellular loop is not a defining feature of TET function, since it is absent in a subgroup of human TETs called the four-transmembrane L6 superfamily (Wright et al., 2000). Of the 17 TET genes in Arabidopsis (Wang et al., 2012), one, TET1/TORNADO2/EKEKO, has been shown to be required for leaf and root patterning (Olmos et al., 2003; Cnops et al., 2006). In addition, like vcc and ops mutant lines, mutants in TET1/TORNADO2/EKEKO also show defects in cotyledon vein patterning (Cnops et al., 2006). Some plant TETs localize to the plasma membrane or to the endoplasmic reticulum when expressed in protoplasts (Boavida et al., 2013). TET3 and a DUF1218-containing protein (AT1G15480) have been found to localize to the plasma membrane and to be enriched in plasmodesmata (Fernandez-Calvino et al., 2011). This raises the interesting possibility that the DUF1218 family also could share functional similarities with TET proteins.
Truernit et al. (2012) reported that mutations in OPS display vascular developmental defects. Mutant plants for OPS developed discontinuous phloem strands in roots and reduced vascular pattern complexity in cotyledons. OPS is expressed in procambial cells and becomes restricted to phloem cells after vascular cell specification. The overexpression of OPS leads to an increase of vascular pattern complexity and premature phloem differentiation, indicating that accelerated phloem development could positively reinforce vascular patterning and lead to accelerated development of both xylem and phloem (Truernit et al., 2012). This suggests that OPS affects both vascular patterning and vascular differentiation and that these are somehow interconnected processes. OPS associates polarly with the plasma membrane of provasculature cells, but its molecular function is unknown. Mutants for VCC and OPS have in common decreased vascular network complexity and the occurrence of disconnected veins in cotyledons. In addition, both genes are expressed in procambial cells, and their overexpression leads to more complex vein networks in cotyledons. The double vcc ops mutant showed stronger defects in cotyledon vascular network complexity than the single mutants (Fig. 6, B–D), suggesting that VCC and OPS have partially overlapping roles in the development of cotyledon vasculature (Fig. 8C).
Interestingly, VCC does not seem to affect auxin transport, suggesting that it is not directly involved in the localization of auxin carriers at the plasma membrane. Although we have not been able to determine the subcellular localization of VCC due to the inability of our GFP fusion construct to rescue the vcc mutant phenotypes, previous proteomic and fluorescent tagging studies have identified the DUF1218-containing proteins encoded by At4g31130 and At3g15480 at the plasma membrane (Hem et al., 2007; Fernandez-Calvino et al., 2011). The fact that VCC and OPS are able to interact physically strongly suggests that VCC and OPS form part of a signaling module at the plasma membrane that controls provasculature specification in embryos. Experiments are under way to test this model.
MATERIALS AND METHODS
Plant Growth and Mutant Isolation
Arabidopsis (Arabidopsis thaliana) seeds were stratified at 4°C for 48 h in the dark and grown for 1 week on plates containing 0.5× Murashige and Skoog medium with 0.6% (w/v) agar and then transferred to soil (23°C under a 16-h-light/8-h-dark cycle). The seed stocks used, SALK_023737C (vcc-1), SALK_047972C (vcc-2), SAIL_237_C09 (vcc-3), SALK_089722C (ops-3), SALK_042563C (ops-4), and pDR5rev:GFP (Friml et al., 2003), were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu). pPIN:PIN1-GFP (Heisler et al., 2005) seeds were kindly donated by Eliot Meyerowitz. Mutant plants were genotyped by PCR using primers 5′-CTCGTTAAGACTTTCCACTACCC-3′ and 5′-ATAGTCAAGAACAAGGACCTACC-3′ to amplify the wild-type VCC allele in vcc-1 and vcc-3 and primers 5′-CTCGTTAAGACTTTCCACTACCC-3′ and 5′-GACAAGGATTCACATAGGTGTG-3′ to amplify the wild-type VCC allele in vcc-2. To amplify vcc mutant alleles, we used 5′-CTCGTTAAGACTTTCCACTACCC-3′ and LBba1 SALK for vcc-1, 5′-GACAAGGATTCACATAGGTGTG-3′ and LBba1 SALK for vcc-2, and 5′-ATAGTCAAGAACAAGGACCTACC-3′ and LB SAIL for vcc-3. pPIN1:PIN1-GFP and pDR5:GFP plants were crossed with vcc-2 plants, and homozygous plants for the vcc-2 insertion and expressing GFP were isolated from F2 plants.
Constructs and Plasmids
pVCC:GUS was obtained by cloning a fragment of approximately 1,900 bp from upstream of the VCC coding sequence (pVCC) between the BamHI and NcoI sites in pCAMBIA1381 using primers 5′-TGGATCCATCCGGAGGTTCACGAATCATGG-3′ and 5′-TCCTATCTTTGCCATGGTCAAATCTCTTAACTTAG-3′.
To make the pVCC:VCC-GFP construct, the vector pCAMBIA1381 was modified by removing the GUS coding region using the PstI and BstEII sites and replacing it with eGFP, which was amplified from pDONR221-eGFP using the forward 5′-CTGCAGATGGTGAGCAAGG-3′ and reverse 5′-GGTCACCTTACTTGTACAGCTCG-3′ primers, containing PstI and BstEII sites, respectively. pVCC:VCC (without stop codon) was cloned upstream of eGFP to allow for translational fusion. pVCC:VCC was amplified from genomic DNA using primers containing the restriction enzymes BamHI and PstI: 5′-GGATCCTGTCTTTGCTAAAGC-3′ and 5′-CTGCAGCTTAGCTTCATCTTTG-3′.
pCaM35S::VCC was obtained by cloning of the VCC genomic sequence between BamHI and SacI in a modified pBI121 in which the GUS coding sequence was removed by digestion with the same restriction enzymes. VCC sequence was amplified from genomic DNA using primers 5′-AAAGGATCCTTACTCGTTAAGACTTTCC-3′ and 5′-CCGGAGCTCTTTGTCACTTAGCTTCATC-3′.
All constructs were sequenced and used to transform Agrobacterium tumefaciens strain GV3101 for transformation into plants.
Protein Alignment and Phylogenetic Tree Calculation
Sequences for the 15 DUF1218-containing proteins were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/), Phytozome (http://www.phytozome.net/), and NCBI (http://www.ncbi.nlm.nih.gov/). Alignment of protein sequences was performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and manual corrections. The alignment was used to build a distance tree using the maximum likelihood method in MEGA5 software (Tamura et al., 2011). The phylogenetic tree was calculated by using the maximum likelihood method based on the Jones-Taylor-Thornton substitution matrices (Jones et al., 1992). The bootstrap consensus tree inferred from 500 replicates was selected to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in fewer than 50% of bootstrap replicates are collapsed. Initial trees for the heuristic search were obtained automatically by applying NJ (Neighbor-Join) and BioNJ algorithms to a matrix of pairwise distances estimated using the Jones-Taylor-Thornton model and then selecting the topology with superior log likelihood value. The analysis involved 63 amino acid sequences. There were a total of 612 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Analysis of Vascular Networks
One-week-old seedlings were clarified in 100% ethanol to remove chlorophyll. The cotyledons were dissected and imaged using either dark-field or epifluorescence analysis with UV light excitation for detecting lignin autofluorescence in an Olympus BX60 microscope (U-MWU Olympus UV excitation cube; excitation, 330–385; beam splitter, 400; emission, 420 long-pass filter). For the analysis of vein patterns in rosette leaves, first leaves from 3-week-old plants grown in soil were transferred into a vial containing 100% ethanol; once clarified, leaves were visualized using a stereoscopic microscope (Nikon SMZ800). Images were taken with a Nikon Coolpix 4500 CCD digital camera.
For histological analysis, seeds from Col-0, vcc-3, ops-4, and vcc-3 ops-4 were rehydrated overnight, and the mature embryos were removed from the seed coat and subjected to pseudo-Schiff propidium iodide staining as described (Truernit et al., 2008). A Zeiss 710 spectral confocal laser scanning microscope was used for imaging. The excitation wavelength was 488 nm for propidium iodide. Data were processed and analyzed using Osirix software (http://www.osirix-viewer.com/).
Iron Staining
Iron staining was performed according to Roschzttardtz et al. (2009). The embryos were dissected from dry seeds previously imbibed in distilled water for 3 h. The isolated embryos were vacuum infiltrated with a solution containing 2% (v/v) HCl and 2% (w/v) potassium ferrocyanide for 15 min and incubated for 30 min at room temperature. After washing with distilled water, the embryos were incubated in a methanol solution containing 10 mm NaN3 and 0.3% (v/v) hydrogen peroxide (H2O2) for 1 h and then washed with 100 mm sodium phosphate buffer (pH 7.4). For the intensification reaction, the embryos were incubated 10 min in 100 mm sodium phosphate buffer (pH 7.4) solution containing 0.025% (w/v) 3,3′-diaminobenzidine tetrahydrochloride hydrate (Sigma-Aldrich), 0.005% (v/v) H2O2, and 0.005% (w/v) CoCl2·2H2O. The reaction was stopped by rinsing with distilled water. The embryos were visualized using a stereoscopic microscope (Nikon SMZ800) and imaged with a Nikon Coolpix 4500 CCD digital camera.
GUS Staining
For histochemical localization of GUS activity, tissues were vacuum infiltrated with 50 mm sodium phosphate buffer (pH 7.4) containing 2 mm each potassium ferrocyanide and potassium ferricyanide, 1% (w/v) Triton X-100, 0.2% (v/v) Tween 20, and 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA cyclohexiammonium salt (Sigma-Aldrich). Incubation was performed in the dark at 37°C for 15 h. Tissues were clarified with ethanol:acetic acid (3:1), washed in 70% (v/v) ethanol, and visualized using a stereoscopic microscope (Nikon SMZ800) equipped with a Nikon Coolpix 4500 CCD digital camera. For anatomical analysis, tissues stained with 5-bromo-4-chloro-3-indolyl-β-d-GlcA cyclohexiammonium salt were dehydrated in successive solutions of 50%, 70%, 90%, and 100% (v/v) ethanol and embedded with Technovit 7100 resin (Kulzer) according to the manufacturer’s instructions. Sections approximately 3 μm in thickness were imaged using an Olympus BX60 microscope.
In Situ Hybridization
In situ hybridization of VCC transcripts on seed sections was performed as described previously (Acevedo et al., 2004) with some modifications. A PCR fragment of 200 bp coding for the C terminus of VCC was cloned in the sense and antisense orientations into pJET (Thermo). Sense or antisense biotin-labeled RNA probes were generated using T7 polymerase and an RNA labeling kit (Epicentre). Formaldehyde-acetic acid-fixed and paraffin-embedded tissue blocks were cut into 10-μm sections and mounted on poly-l-Lys-coated glass slides (Lab Scientific). After removal of paraffin in xylene and ethanol, the slides were immersed in diethylpyrocarbonate-treated double distilled water. The sections were treated with 0.1 m sodium citrate buffer (pH 6.2) as target retrieval solution at 95°C for 20 min, cooled at room temperature, and washed with diethylpyrocarbonate-treated double distilled water. They were treated with pepsin (Sigma-Aldrich) for 15 min at 37°C. Pepsin was washed and inactivated by immersing the sections twice in 0.2% (w/v) Gly phosphate-buffered saline (pH 7.6) for 3 min and in diethylpyrocarbonate-treated double distilled water for 3 min. Sections were then immersed in 100% ethanol for 2 min, air dried, prehybridized in hybridization buffer (50% [v/v] formamide, 1× SSC, 1× Denhardt’s solution [Amresco], 50 mm EDTA, 500 μg mL−1 tRNA, and 50 mm HEPES, pH 7) at 37°C for 30 min, and then hybridized with sense or antisense RNA probes overnight at 37°C in hybridization buffer. Sections were then washed once in 0.1× SSC (3 m NaCl and 0.3 m sodium citrate, pH 7) and twice stringently with 0.1× SSC for 15 min at 42°C before treatment with 0.3% (v/v) H2O2 in methanol for 30 min to block endogenous peroxidase activity. For signal detection, a 1:500-fold diluted primary streptavidin-horseradish peroxidase (0.5 mg mL−1; Thermo Scientific) solution was applied for 30 min on each section at room temperature. The sections were washed three times with phosphate-buffered saline-0.05% Tween 20 (v/v) for 5 min. Signals were developed with metal-enhanced diaminobenzidine (DAB; Thermo Scientific) chromogen solution.
Split-Ubiquitin Yeast Two-Hybrid Assay
The split-ubiquitin yeast two-hybrid assay was performed using the DUAL membrane starter kit (Dualsystems Biotech). Complementary DNAs (cDNAs) were amplified by RT-PCR from Col-0 plants and cloned directly in pPR3-N prey/pBT3-C bait vectors by SfiI restriction sites included in the primers; plasmids were transformed into Escherichia coli and sequenced. The yeast (Saccharomyces cerevisiae) strain used was NMY51 (Dualsystems Biotech). Yeast cells were cotransformed by thermic shock with pBT3-C-OPS and pPR3-N-VCC, pBT3-C and pPR3-N VCC, or pBT3-C-OPS and pPR3-N. The number of colonies grown on -LWH medium containing 15 mm 3-AT (selection for interaction) was divided by the number of colonies grown on -LW medium (selection for transformation) to calculate the percentage of colonies showing a VCC-OPS positive interaction. Between 500 and 3,000 colonies were counted in each case.
Bimolecular Fluorescence Complementation Assay
VCC and OPS coding regions were cloned into the split YFP vectors pSY735 and pSY736 (Bracha-Drori et al., 2004). Both vectors were used to transfect protoplasts from 30-d-old plants as described previously (Yoo et al., 2007). Approximately 2 × 105 protoplasts were transfected with 1 µg of each vector and then incubated for 16 h. Empty vectors were used as negative controls, and a vector containing a pCaMV35S:CFP (pAVA574) reporter (von Arnim et al., 1998) was cotransfected to assess transfection efficiency. To analyze interactions, YFP and cyan fluorescent protein (CFP) fluorescence were detected using a Zeiss LSM 510 META confocal laser scanning microscope and a 20× numerical aperture 0.5 objective. YFP was excited with an argon laser at 514 nm, and the emission was collected using a band pass 535- to 590-nm infrared-blocking filter. CFP was excited with an argon laser at 458 nm, and the emission was collected using a band pass 480 to 520 nm, infrared-blocking filter. A total of 200 protoplasts for each vector combination were scored for CFP fluorescence and reconstitution of YFP fluorescence. The experiment was repeated twice.
Confocal Microscopy
Isolated embryos from vcc-2/pPIN1:PIN1-GFP and vcc-2/pDR5rev:GFP plants were visualized using a Zeiss 510 Meta confocal laser scanning microscope. GFP was excited with 488 nm, and emission was collected with a band pass 500 to 530 nm, infrared-blocking filter. Images were edited using the LSM image browser (http://www.zeiss.com/lsm) and Adobe Photoshop CS4.
RT-PCR and RT-Quantitative PCR Analyses
For RT-PCR, total RNA from plant tissues (leaves, inflorescences, torpedo/bent cotyledon embryos, and seedlings) was isolated using the RNeasy Mini Kit (Qiagen). cDNA was obtained using 1 to 2 μg of RNA, oligo(dT), and Avian Myeloblastosis Virus reverse transcriptase (Promega) in a 20-μL reaction volume. Then, 1 to 3 μL of RT product was used as template in a 25-μL volume reaction, and different PCR cycles were performed for each transcript: 35 or 25 cycles (30 s at 95°C, 30 s at 55°C, and 1 min at 72°C) for VCC, OPS, BANYULS (BAN), and SUCCINATE DEHYDROGENASE2-3 (SDH2-3) and 30 cycles (30 s at 95°C, 30 s at 55°C, and 1.5 min at 72°C) for ACTIN2 and PROTEIN PHOSPHATASE2A (PP2A). The PCRs were performed using EconoTaq PLUS 2X master mix (Lucigen). Primers used were as follows: 5′-CTCGTTAAGACTTTCCACTACCC-3′ and 5′-ATAGTCAAGAACAAGGACCTACC-3′ for VCC; 5′-TTTGTTGATCAATACAGCCTCATTACAC-3′ and 5′-GCGACGAGATTGTTGAAGTTAGAG-3′ for OPS; 5′-TTCCGCTCTTTCTTTCCAAGCTCA-3′ and 5′-AAGAGGCATCAATTCGATCACTCA-3′ for ACTIN2; 5′-CGTCTGTCTTGCGGTTGTTGGG-3′ and 5′-CACTCTCTGTTCTCACTAACGGATC-3′ for SDH2-3; 5′-GGACCAGACTCTTACACACACCGG-3′ and 5′-AAGCCCTCTTCGAATTCTGACAACAC-3′ for BAN; and 5′-TAACGTGGCCAAAATGATGC-3′ and 5′-GTTCTCCACAACCGCTTGGT-3′ for PP2A.
For the RT-quantitative PCR analysis, total RNA was extracted from 7-d-old seedlings, roots from 10-d-old plants, leaves from 4-week-old plants, flowers and inflorescences from 5- to 7-week-old plants, and inflorescence stems (two nodes from the base of the stem), hypocotyls, and siliques from 2-month-old plants using the RNeasy Plant Mini Kit (Qiagen). Dry seed RNA was prepared by using the protocol described by Oñate-Sánchez and Vicente-Carbajosa (2008). RNA samples were treated with RQ1 DNase (Promega) according to the manufacturer’s instructions. Three biological replicates for each tissue type were used. First-strand cDNA synthesis (RT) reaction and PCR were done in the same tube using the qScript One-Step qRT-PCR Kit (Quanta Biosciences) using the primers VCC1-qRT-F (5′-CGTTCAGGCTCGGTCTAGGT-3′), VCC1-qRT-R (5′-AGCGAATACGATCCAAGTGAG-3′), PP2A-F (5′-TAACGTGGCCAAAATGATGC-3′), and PP2A-R (5′-GTTCTCCACAACCGCTTGGT-3′). Four technical replicates per sample and several negative controls with no reverse transcriptase were included. Samples were run on a LightCycler 480 Real Time-PCR System (Roche Applied Science), and LinRegPCR was used to analyze the data (Ramakers et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ACTIN2 (AT3G18780), BAN (AT1G61720), OPS (AT3G09070), PP2A (AT1G69960), SDH2-3 (AT5G65165), and VCC (AT2G32280).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular phylogenetic analysis based on maximum likelihood.
Supplemental Figure S2. vcc embryos show abnormal cotyledon vein complexity.
Supplemental Figure S3. PIN1-GFP localization in control (A and B) and vcc mutant (C and D) embryos.
Supplemental Figure S4. Expression pattern of VCC.
Supplemental Figure S5. Expression of VCC in root and embryo.
Supplemental Table S1. Quantitative analysis of vascular network complexity and connectivity in several lines shown in Figures 2 and 6.
Supplemental Table S2. Quantitative analysis (in percentages) of vascular network complexity and connectivity in control and VCC-overexpressing lines.
Supplementary Material
Acknowledgments
We thank Bindiya Sahah and Teresa Thayyil (University of Wisconsin-Madison) for their assistance in preparing plant material for this project, Julian Verdonk (University of Wisconsin-Madison) for the initial identification of the DUF1218 gene family, and Dr. Sarah Swanson (Newcomb Imaging Center, University of Wisconsin, Madison) for assistance with confocal imaging.
Glossary
- RLK
receptor-like kinase
- NCBI
National Center for Biotechnology Information
- T-DNA
transfer DNA
- RT
reverse transcription
- Col-0
Columbia-0
- 3-AT
3-aminotriazole
- YFP
yellow fluorescent protein
- H2O2
hydrogen peroxide
- DAB
diaminobenzidine
- cDNA
complementary DNA
- CFP
cyan fluorescent protein
Footnotes
This work was supported by the Department of Energy Great Lakes Bioenergy Research Center (grant no. DE–FC02–07ER64494), the National Science Foundation (grant no. MCB1157824), and the University of Wisconsin Graduate School (grant to M.S.O.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Acevedo FG, Gamboa A, Paéz-Valencia J, Jiménez-García LF, Izaguirre-Sierra M, Alvarez-Buylla ER. (2004) FLOR1, a putative interaction partner of the floral homeotic protein AGAMOUS, is a plant-specific intracellular LRR. Plant Sci 167: 225–231 [Google Scholar]
- Agusti J, Lichtenberger R, Schwarz M, Nehlin L, Greb T. (2011) Characterization of transcriptome remodeling during cambium formation identifies MOL1 and RUL1 as opposing regulators of secondary growth. PLoS Genet 7: e1001312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Qin P, Broz M, Becker JD, McCormick S. (2013) Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol 163: 696–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Bryan AC, Obaidi A, Wierzba M, Tax FE. (2012) XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235: 111–122 [DOI] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Nelson T. (2010) The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceserani T, Trofka A, Gandotra N, Nelson T. (2009) VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J 57: 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, Malenica N, Luschnig C, Tietz O, Ditengou F, Palme K, et al. (2006) The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18: 852–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Cordner G, Beebe D, Sieburth LE. (2000) The SCARFACE gene is required for cotyledon and leaf vein patterning. Development 127: 3205–3213 [DOI] [PubMed] [Google Scholar]
- Dhondt S, Van Haerenborgh D, Van Cauwenbergh C, Merks RMH, Philips W, Beemster GTS, Inzé D. (2012) Quantitative analysis of venation patterns of Arabidopsis leaves by supervised image analysis. Plant J 69: 553–563 [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E. (2009) Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246 [DOI] [PubMed] [Google Scholar]
- Eisses JF, Kaplan JH. (2005) The mechanism of copper uptake mediated by human CTR1: a mutational analysis. J Biol Chem 280: 37159–37168 [DOI] [PubMed] [Google Scholar]
- Elo A, Immanen J, Nieminen K, Helariutta Y. (2009) Stem cell function during plant vascular development. Semin Cell Dev Biol 20: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Etchells JP, Provost CM, Mishra L, Turner SR. (2013) WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140: 2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Turner SR. (2010) The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A. (2011) Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Turner S. (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hejátko J, Ryu H, Kim GT, Dobesová R, Choi S, Choi SM, Soucek P, Horák J, Pekárová B, Palme K, et al. (2009) The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21: 2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hem S, Rofidal V, Sommerer N, Rossignol M. (2007) Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem Biophys Res Commun 363: 375–380 [DOI] [PubMed] [Google Scholar]
- Hemler ME. (2005) Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6: 801–811 [DOI] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M. (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Hou H, Erickson J, Meservy J, Schultz EA. (2010) FORKED1 encodes a PH domain protein that is required for PIN1 localization in developing leaf veins. Plant J 63: 960–973 [DOI] [PubMed] [Google Scholar]
- Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI. (2009) Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA 106: 13630–13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M. (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Miyashima S, Sebastian J, Lee JY, Helariutta Y. (2013) Stem cell function during plant vascular development. EMBO J 32: 178–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Yadegari R, Tax FE. (2007) RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev Cell 12: 943–956 [DOI] [PubMed] [Google Scholar]
- Nose Y, Rees EM, Thiele DJ. (2006) Structure of the Ctr1 copper trans‘PORE’ter reveals novel architecture. Trends Biochem Sci 31: 604–607 [DOI] [PubMed] [Google Scholar]
- Olmos E, Reiss B, Dekker K. (2003) The ekeko mutant demonstrates a role for tetraspanin-like protein in plant development. Biochem Biophys Res Commun 310: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM. (2008) Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J 56: 251–263 [DOI] [PubMed] [Google Scholar]
- Pullen M, Clark N, Zarinkamar F, Topping J, Lindsey K. (2010) Analysis of vascular development in the hydra sterol biosynthetic mutants of Arabidopsis. PLoS ONE 5: e12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S. (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S. (2010) Straightforward histochemical staining of Fe by the adaptation of an old-school technique: identification of the endodermal vacuole as the site of Fe storage in Arabidopsis embryos. Plant Signal Behav 5: 56–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. (2000) Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol 41: 649–656 [DOI] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wisniewska J, Reinöhl V, Friml J, Benková E. (2006) Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20: 2902–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Helariutta Y. (2010) Vascular pattern formation in plants. Curr Top Dev Biol 91: 221–265 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T. (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. (1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Sieburth LE. (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Deyholos MK. (2006) Vascular development: the long and winding road. Curr Opin Plant Biol 9: 48–54 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM. (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter M, Topping J, Pullen M, Friml J, Palme K, Hackett R, Grierson D, Lindsey K. (2002) hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp CS, Kolesnikova TV, Hemler ME. (2003) Functional domains in tetraspanin proteins. Trends Biochem Sci 28: 106–112 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Belcram K, Barthélémy J, Palauqui J-C. (2012) OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139: 1306–1315 [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC. (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Wang F, Vandepoele K, Van Lijsebettens M. (2012) Tetraspanin genes in plants. Plant Sci 190: 9–15 [DOI] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA. (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci USA 107: 22338–22343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B. (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–62512615936 [Google Scholar]
- Wright MD, Ni J, Rudy GB. (2000) The L6 membrane proteins: a new four-transmembrane superfamily. Protein Sci 9: 1594–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Richter GL, Wang X, Młodzińska E, Carraro N, Ma G, Jenness M, Chao DY, Peer WA, Murphy AS. (2013) Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74: 37–47 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang J, Elo A, Helariutta Y. (2011) Arabidopsis as a model for wood formation. Curr Opin Biotechnol 22: 293–299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.