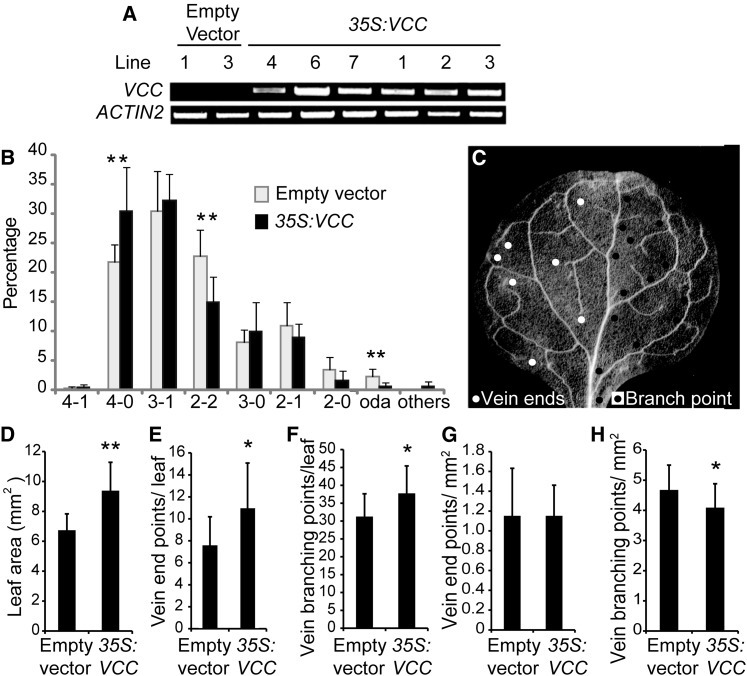
Figure 3.
VCC overexpression increases the complexity of vein networks in cotyledons and first leaves. A, RT-PCR analysis of VCC expression in Col-0 plants transformed with either an empty vector or the pCaMV35S:VCC construct (35S:VCC). cDNA was obtained using 1 μg of RNA, and 2 μL of RT product was used as a template in a 25-μL volume reaction. Twenty-five PCR cycles were performed to amplify VCC. ACTIN2 was used as a control. B, Distribution of vein complexity patterns in pCaMV35S:VCC and plants transformed with an empty vector. T2 embryos from seven T1 lines transformed with an empty vector and nine T1 pCaMV35S:VCC lines were analyzed. For each line, between 100 and 200 individual cotyledons were examined. C, Dark-field image of a first rosette leaf from a 3-week-old Arabidopsis plant. For clarity, vein end points (white circles at left side) and vein branching points (black circles at right side) considered for vein complexity analysis are indicated in only half of the leaf blade. D, Quantitative analysis of first leaf area. E, Quantification of vein end points and branching points per leaf. F, Vein branching points per leaf. G, Ratio between vein end points and leaf blade area. H, Ratio between vein branching points and leaf blade area. In total, 12 and 28 first rosette leaves from T1 plants transformed with the empty vector and the pCaMV35S:VCC construct, respectively, were analyzed. For Student’s t test analysis, P < 0.05 (*) and P < 0.01 (**).